AbstractThe complete chloroplast genome of Zoysia macrostachya Franch. & Sav. isolated in Korea is 135,902 bp long (GC ratio is 38.4%) and has four subregions; 81,546 bp of large single-copy (36.3%) and 12,586 bp of small single-copy (32.7%) regions are separated by 20,885 bp of inverted repeat (44.1%) regions, including 130 genes (83 protein-coding genes, eight rRNAs, and 39 tRNAs). Thirty-nine single nucleotide polymorphisms and 11 insertions and deletion (INDEL) regions were identified from two Z. macrostachya chloroplast genomes, the smallest among other Zoysia species. Phylogenetic trees show that two Z. macrostachya chloroplast genomes are clustered into a single clade. However, we found some incongruency with regard to the phylogenetic position of the Z. macrostachya clade. Our chloroplast genome provides insights into intraspecific variations and species delimitation issues pertaining to the Zoysia species.
Zoysia macrostachya Steud. is a perennial plant species, majorly distributed in Korea, Japan, and east costal of China (Soreng et al., 2015; Park et al., 2020a). Zoysia macrostachya is a warm season grass having C-4 photosynthetic system (Moser et al., 2004), which can grow well under the high temperature. Till now, no commercial cultivars of Z. macrostachya has been developed (Loch et al., 2017; Wang et al., 2020), which is different from Zoysia japonica (Chai and Kim, 2000; Ge et al., 2006; Sun et al., 2010) and Zoysia matrella (Bae et al., 2008; Choi et al., 2017). Because Z. macrostachva has more resistance to salt than Z. japonica and Z. matrella (Loch et al., 2017), it is highly valuable as a breeding copy for land reclamation or coastal areas. Due to recently sequenced chloroplast genomes of Zoysia (Tanaka et al., 2016; Lee and Park, 2021a; Lee and Park, 2021b), intraspecific variations which have been utilized for developing markers (Li et al., 2020) and understanding phylogenetic relationship (Park et al., 2020b; Park et al., 2021a) can be investigated along with Zoysia species. Here, we completed the complete chloroplast genome of Z. macrostachya to understand intraspecific variations as well as species boundary.
Materials and MethodsPlant materialWe isolated the Z. macrostachya in the Subtropical Horticulture Research Institute, Jeju city, Korea (36.83914N, 127.17096E) for conserving natural isolate in Jejudo island in Korea. A voucher and isolated DNA was deposited in the InfoBoss Cyber Herbarium (IN, the voucher number IB-01097).
DNA extraction and mitochondrial genome determinationIts total DNA was extracted from fresh leaf by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using NovaSeq6000 at Macrogen Inc., Korea, and de novo assembly was done by Velvet v1.2.10 (Zerbino and Birney, 2008) and GapCloser v1.12 (Zhao et al., 2011). Assembled sequences were modified and confirmed by BWA v0.7.17 (Li, 2013) and SAMtools v1.9 (Li et al., 2009). Circular form was confirmed by connecting both ends using GapCloser v1.12. All analyses were conducted in the Genome Information System (http://geis.infoboss.co.kr/) used in the previous studies (Kim et al., 2021a, 2021b; Park et al., 2019a, 2019d, 2021f).
Genome annotation was conducted based on the Z. macrostachya chloroplast reported previously (NC_042189) (Cheon et al., 2021) with Geneious Prime 2020.2.4 (Biomatters Ltd, Auckland, New Zealand). A circular map of Z. macrostachya chloroplast genome was drawn using OGDRAW v1.31 (Greiner et al., 2019).
Identification of intraspecific variationsSingle nucleotide polymorphisms (SNPs) and insertions and deletions (INDELs) were identified from the pairwise sequence alignment of the two Z. macrostachya chloroplast genomes conducted by MAFFT 7.450 (Katoh and Standley, 2013) with ‘Find variations/SNPs’ implemented in Geneious Prime 2020.2.4 (Biomatters Ltd., Auckland, New Zealand), which has been used in the previous studies investigating intraspecific variations on organelle genomes (Park et al., 2020c, 2021b, 2021e). INDEL region was defined as the continuous INDELs.
Phylogenetic analysisMaximum-Likelihood (ML), Neighbor-Joining (NJ), and Bayesian inference (BI) phylogenetic trees were constructed based on the multiple sequence alignment of ten Zoysia chloroplast genomes by MAFFT v7.450 (Katoh and Standley, 2013). The NJ and ML tree were reconstructed in MEGA X with 10,000 and 1,000 bootstrap repeats, respectively (Kumar et al., 2018). In the ML analysis, a heuristic search was used with nearest-neighbor interchange branch swapping, TVM + F + R4 model, and uniform rates among sites. All other options used the default settings. The posterior probability of each node was estimated by BI using MrBayes v3.2.6 (Ronquist et al., 2012) plug-in implemented in Geneious Prime 2020.2.4 (Biomatters Ltd., Auckland, New Zealand). The HKY85 model with gamma rates was used as a molecular model. A Markov chain Monte Carlo algorithm was employed for 1,100,000 generations, sampling trees every 200 generations, with four chains running simultaneously. Trees from the first 100,000 generations were discarded as burn-in.
Data Availability StatementChloroplast genome sequence can be accessed via accession number MZ233426 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA730583, SAMN19236164, and SRR14572437, respectively.
Results and DiscussionThe chloroplast genome of Z. macrostachya (GenBank accession no. MZ233426) is 135,902 bp (GC ratio, 38.4%) and has four subregions: 81,546 bp of large single copy (LSC; 36.3%) and 12,586 bp of small single copy (SSC; 32.7%) regions are separated by 20,885 bp of inverted repeat (IR; 44.1%) (Fig. 1). Lengths of the complete Zoysia chloroplast genomes displayed 135,810 bp to 135,904 bp, similar to those of Tritichum, Oryza, and Avena genera (Ogihara et al., 2000; Wambugu et al., 2015; Liu et al., 2020) and displayed the short length among available Poaceae chloroplast genomes (from 129,905 bp [Rytidosperma semiannulare (Labill.) Connor & Edgar; NC_036701] to 162,086 bp [Paspalum ionanthum Chase; NC_039464] (Burke et al., 2018)). It contains 130 genes (83 protein-coding genes, eight rRNAs, 39 tRNAs, and one pseudogene); 19 genes (seven protein-coding genes, four rRNAs, and eight tRNAs) are duplicated in inverted repeat regions (Fig. 1). One pseudogene is a partial ndhF, same to that of the previously reported chloroplast genome of Z. macrostachya (Cheon et al., 2021).
Thirty-nine SNPs and 11 INDEL regions (18 bp in total) were identified from pairwise alignment of two Z. macrostachya chloroplast genomes. The longest INDEL region is 6 bp long and two INDEL regions are 2-bp. Number of SNPs identified in Z. macrostachya is greater in number than that of Z. matrella (28 SNPs) (Lee and Park, 2021a); while number of the INDEL regions is smaller. In addition, both are smaller than those of Z. japonica (68 SNPs and 24 INDEL regions) (Lee and Park, 2021b), indicating that intraspecific variations of Z. macrostachya is the smallest among Zoysia species. These numbers are also smaller than those identified between the samples isolated in Korea: e.g., Campanula takesimana (Park et al., 2021a), Pseudostellaria palibiniana (Kim et al., 2019), Daphne genkwa (Yoo et al., 2021), Abeliophyllum distichum (Min et al., 2019; Park et al., 2019b, 2019c, 2021d), Chenopodium album (Park et al., 2021c), and Pyrus ussuriensis (Cho et al., 2019).
Two and one non-synonymous SNPs were found in psaB and psbK, respectively; while two, one, and one synonymous SNPs were identified in rpoC2, infA, and ccsA, respectively. In addition, one SNP was in trnQ, displaying 20.51% SNPs are in genic region. Number of genes containing SNPs in Z. macrostachya is smaller than that of Z. japonica, exhibiting different genes: rpoB, rpoC2, atpB, and ndhA contain one synonymous SNP each, petA covers three synonymous SNPs, rpoC1 and atpF have one non-synonymous SNP each (Lee and Park, 2021b). This distribution of intraspecific SNPs in the genic region can be developed as intraspecific molecular markers with additional experiments of validation. In addition, ycf1 which exhibits high nucleotide diversity in various plant species (Jiang et al., 2017; Park and Oh, 2020; Loeuille et al., 2021) was not found in this chloroplast genome, congruent to the other Zoysia chloroplast genomes (Tanaka et al., 2016; Cheon et al., 2021; Lee and Park, 2021a, 2021b).
ML and BI phylogenetic trees show that our Z. macrostachya was clustered with previous Z. macrostachya with high supportive values of all trees (Fig. 1). Interestingly, the branch length in Z. macrostachya clade is similar to the clade containing Z. japonica and Z. sinica (Fig. 2), suggesting the possible scenario that Z. sinica can be considered as Z. japonica together with similar morphological features of the two species (Yu et al., 1974). In addition, phylogenetic position of the Z. macrostachya clade was not supported by the BI tree (Fig. 2) and topology of Z. macrostachya and Z. macrantha presented in Cheon et al. (2021) was not also congruent with the three trees (Fig. 2), suggesting additional chloroplast genomes of neighbor species of Z. macrostachya are required to clarify the phylogenetic relationship. Taken together, our chloroplast genome provides the insight of intraspecific variations and species delimitation of Zoysia species.
ACKNOWLEDGMENTSThis work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ014235)” and “Next-Generation BioGreen 21 Program (Project No. PJ 01368504)” Rural Development Administration, Republic of Korea.
Fig. 1.Circular map of chloroplast genome of Zoysia macrostachya isolated in Korea. Gene shown outside are transcribed clockwise, and inside the circle are transcribed counter clockwise. Genes are color-coded to distinguish different functional groups. The dark grey and the light grey plot in the inner circle correspond to the GC content and AT content, respectively. 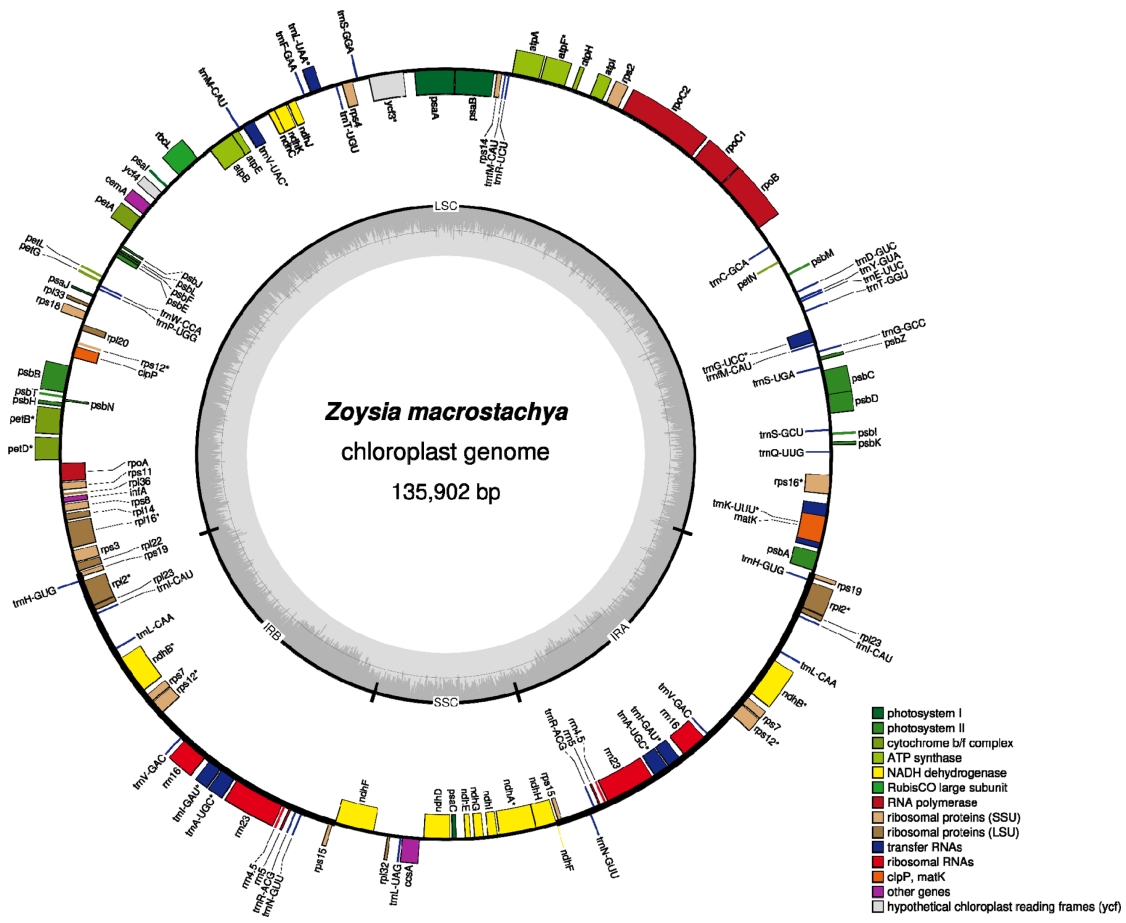
Fig. 2.Maximum-Likelihood (ML), Neighbor-Joining (NJ), and Bayesian inference (BI) phylogenetic trees of ten Zoysia and one outgroup complete chloroplast genomes. Phylogenetic tree was drawn based on ML tree. The numbers and hyphens above branches indicate support values of ML, NJ, and BI trees, and incongruent node in comparison to that of ML tree, respectively. BI tree was displayed separately without outgroup species which displayed too long branch length with blue colored shaded dotted rectangle. 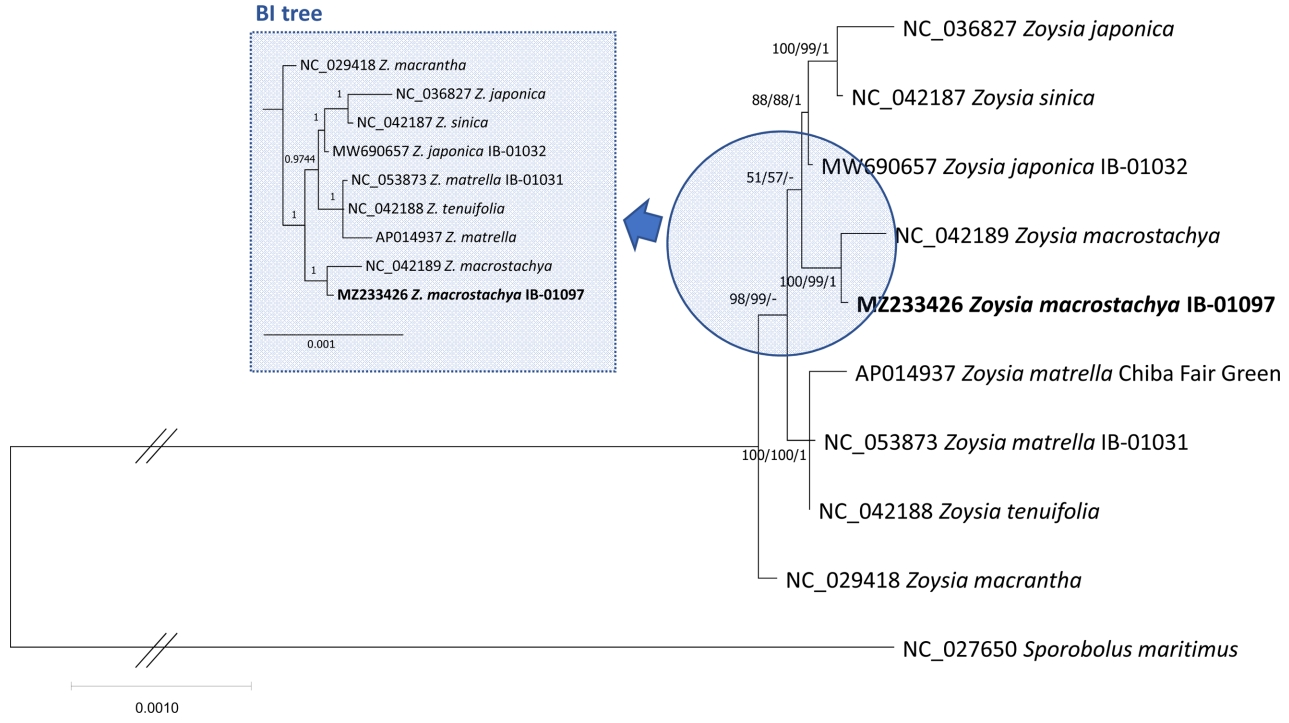
Literature CitedBae, TW. Vanjildorj, E. Song, SY. Nishiguchi, S. Yang, SS. Song, IJ. Chandrasekhar, T. Kang, TW. Kim, JI. Koh, YJ. Park, SY. Lee, J. Lee, Y.-E. Ryu, KH. Riu, KZ. Song, P.-S and Lee, H. Y. 2008. Environmental risk assessment of genetically engineered herbicide-tolerantZoysia japonica
. Journal of Environmental Quality 37: 207-218.
Burke, SV. Ungerer, MC and Duvall, MR. 2018. Investigation of mitochondrial-derived plastome sequences in the Paspalum lineage (Panicoideae; Poaceae). BMC Plant Biology 18: 152.
Chai, M and Kim, D. 2000.
Agrobacterium-mediated transformation of Korean lawngrass (Zoysia japonica). Journal of the Korean Society for Horticultural Science 41: 455-458.
Cheon, S.-H. Woo, M-A. Jo, S. Kim, Y.-K and Kim, K.-J. 2021. The chloroplast phylogenomics and systematics of Zoysia (Poaceae). Plants 10: 1517.
Cho, M-S. Kim, Y. Kim, S-C and Park, J. 2019. The complete chloroplast genome of Korean Pyrus ussuriensis Maxim. (Rosaceae): Providing genetic background of two types of P. ussuriensis
. Mitochondrial DNA Part B 4: 2424-2425.
Choi, J-S. Yang, G-M. Bae, E-J. Park, Y-B and Lee, K-S. 2017. Development of new hybrid zoysiagrass cultivar ‘Seah’. Weed and Turfgrass Science 6: 306-312.
Ge, Y. Norton, T and Wang, Z-Y. 2006. Transgenic zoysiagrass (Zoysia japonica) plants obtained by Agrobacterium-mediated transformation. Plant Cell Reports 25: 792-798.
Greiner, S. Lehwark, P and Bock, R. 2019. OrganellarGenome-DRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Research 47: W59-W64.
Jiang, D. Zhao, Z. Zhang, T. Zhong, W. Liu, C. Yuan, Q and Huang, L. 2017. The chloroplast genome sequence of Scutellaria baicalensis provides insight into intraspecific and interspecific chloroplast genome diversity in Scutellaria
. Genes 8: 227.
Katoh, K and Standley, DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30: 772-780.
Kim, M. Xi, H and Park, J. 2021a. Genome-wide comparative analyses of GATA transcription factors among 19 Arabidopsis ecotype genomes: Intraspecific characteristics of GATA transcription factors. PLoS One 16: e0252181.
Kim, M. Xi, H. Park, S. Yun, Y and Park, J. 2021b. Genome-wide comparative analyses of GATA transcription factors among seven Populus genomes. Scientific Reports 11: 16578.
Kim, Y. Heo, K.-I and Park, J. 2019. The second complete chloroplast genome sequence of Pseudostellaria palibiniana (Takeda) Ohwi (Caryophyllaceae): Intraspecies variations based on geographical distribution. Mitochondrial DNA Part B 4: 1310-1311.
Kumar, S. Stecher, G. Li, M. Knyaz, C and Tamura, K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35: 1547-1549.
Lee, B and Park, J. 2021a. The complete chloroplast genome of Zoysia matrella (L.) Merr. isolated in Korea (Poaceae): Investigation of intraspecific variations on chloroplast genomes. Mitochondrial DNA Part B Resources 6: 572-574.
Lee, B and Park, J. 2021b. The complete chloroplast genome of Zoysia japonica Steud. isolated in Korea (Poaceae): Investigation of potential molecular markers on Z. japonica chloroplast genomes. Plant Biotechnology Report. Advanced online publication. https://doi.org/10.1007/s11816-021-00708-y.
Li, C. Zheng, Y and Huang, P. 2020. Molecular markers from the chloroplast genome of rose provide a complementary tool for variety discrimination and profiling. Scientific Reports 10: 12188.
Li, H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at: https://arxiv.org/abs/1303.3997
.
Li, H. Handsaker, B. Wysoker, A. Fennel, T. Ruan, J. Homer, N. Marth, G. Abecasis, G and Durbin, R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078-2079.
Liu, Q. Li, X. Li, M. Xu, W. Schwarzacher, T and Heslop-Harrison, JS. 2020. Comparative chloroplast genome analyses of Avena: Insights into evolutionary dynamics and phylogeny. BMC Plant Biology 20: 406.
Loch, DS. Ebina, M. Choi, JS and Han, L. 2017. Ecological implications of Zoysia species, distribution, and adaptation for management and use of zoysiagrasses. International Turfgrass Society Research Journal 13: 11-25.
Loeuille, B. Thode, V. Siniscalchi, C. Andrade, S. Rossi, M and Pirani, JR. 2021. Extremely low nucleotide diversity among thirty-six new chloroplast genome sequences from Aldama (Heliantheae, Asteraceae) and comparative chloroplast genomics analyses with closely related genera. PeerJ 9: e10886.
Min, J. Kim, Y. Xi, H. Jang, T. Kim, G. Park, J and Park, J.-H. 2019. The complete chloroplast genome of a new candidate cultivar, Sang Jae, of Abeliophyllum distichum Nakai (Oleaceae): Initial step of A. distichum intraspecies variations atlas. Mitochondrial DNA Part B Resources 4: 3176-3718.
Moser, LE. Burson, BL and Sollenberger, LE. 2004. Warm-Season (C4) Grasses. American Society of Agronomy, Madison, WI. 1171 pp.
Ogihara, Y. Isono, K. Kojima, T. Endo, A. Hanaoka, M. Shiina, T. Terachi, T. Utsugi, S. Murata, M. Mori, N. Takumi, S. Ikeo, K. Gojobori, T. Murai, R. Murai, K. Matsuoka, Y. Ohnishi, Y. Tajiri, H and Tsunewaki, K. 2000. Chinese spring wheat (Triticum aestivum L.) chloroplast genome: Complete sequence and contig clones. Plant Molecular Biology Reporter 18: 243-253.
Park, J. An, J-H. Kim, Y. Kim, D. Yang, B-G and Kim, T. 2020a. Database of National Species List of Korea: The taxonomical systematics platform for managing scientific names of Korean native species. Journal of Species Research 9: 233-246.
Park, J. Bae, Y. Kim, B-Y. Nam, G-H. Park, J-M. Lee, BY. Suh, H- J and Oh, S-H. 2021a. The complete chloroplast genome of Campanula takesimana Nakai from Dokdo Island in Korea (Campanulaceae). Mitochondrial DNA Part B Resources 6: 135-137.
Park, J. Heo, K-I. Kim, Y and Kwon, W. 2019a. The complete chloroplast genome of Potentilla freyniana Bornm. (Rosaceae). Mitochondrial DNA Part B Resources 4: 2420-2421.
Park, J. Kim, Y. Kwon, W. Xi, H and Park, C-H. 2021b. The complete chloroplast genome sequence of new species candidate of Plantago depressa Willd. in Korea (Plantaginaceae). Mitochondrial DNA Part B Resources 6: 1961-1963.
Park, J. Kim, Y. Xi, H. Jang, T and Park, J-H. 2019b. The complete chloroplast genome of Abeliophyllum distichum Nakai (Oleaceae), cultivar Ok Hwang 1ho: Insights of cultivar specific variations of A. distichum
. Mitochondrial DNA Part B Resources 4: 1640-1642.
Park, J. Min, J. Kim, Y and Chung, Y. 2021c. The comparative analyses of six complete chloroplast genomes of morphologically diverse Chenopodium album L. (Amaranthaceae) collected in Korea. International Journal of Genomics 2021: 6643444.
Park, J. Min, J. Kim, Y. Xi, H. Kwon, W. Jang, T. Kim, G and Park, J- H. 2019c. The complete chloroplast genome of a new candidate cultivar, Dae Ryun, of Abeliophyllum distichum Nakai (Oleaceae). Mitochondrial DNA Part B Resources 4: 3713-3715.
Park, J. Park, S. Jang, T. Kim, G and Park, J-H. 2021d. The complete chloroplast genome of Abeliophyllum distichum f. lilacinum Nakai (Oleaceae) from the Chungbuk Province. Mitochondrial DNA Part B Resources 6: 1754-1756.
Park, J. Xi, H and Oh, S-H. 2020. Comparative chloroplast genomics and phylogenetic analysis of the Viburnum dilatatum complex (Adoxaceae) in Korea. Korean Journal of Plant Taxonomy 50: 8-16.
Park, J. Xi, H and Kim, Y. 2021e. The complete mitochondrial genome of Arabidopsis thaliana (Brassicaceae) isolated in Korea. Korean Journal of Plant Taxonomy 51: 176-180.
Park, J. Xi, H and Kim, Y. 2020b. The complete chloroplast genome of Arabidopsis thaliana isolated in Korea (Brassicaceae): An investigation of intraspecific variations of the chloroplast genome of Korean A. thaliana
. International Journal of Genomics 2020: 3236461.
Park, J. Xi, H. Kim, Y. Nam, S and Heo, K-I. 2020c. The complete mitochondrial genome of new species candidate of Rosa rugosa (Rosaceae). Mitochondrial DNA Part B Resources 5: 3435-3437.
Park, J. Xi, H. Son, J. Shin, HT. Kang, H and Park, S. 2021f. The complete chloroplast genome of Castanopsis sieboldii (Makino) Hatus (Fagaceae). Mitochondrial DNA Part B Resources 6: 2743-2745.
Park, J. Yun, N and Oh, S-H. 2019d. The complete chloroplast genome of an endangered species in Korea, Halenia corniculata (L.) Cornaz (Gentianaceae). Mitochondrial DNA Part B Resources 4: 1539-1540.
Ronquist, F. Teslenko, M. Van Der Mark, P. Ayres, DL. Darling, A. Höhna, S. Larget, B. Liu, L. Suchard, MA and Huelsenbeck, JP. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539-542.
Soreng, RJ. Peterson, PM. Romaschenko, K. Davidse, G. Zuloaga, FO. Judziewicz, EJ. Filgueiras, TS. Davis, JI and Morrone, O. 2015. A worldwide phylogenetic classification of the Poaceae (Gramineae). Journal of Systematics and Evolution 53: 117-137.
Sun, H-J. Song, I-J. Bae, T-W and Lee, H-Y. 2010. Recent developments in biotechnological improvement of Zoysia japonica Steud. Journal of Plant Biotechnology 37: 400-407.
Tanaka, H. Hirakawa, H. Muguerza, M. Hashiguchi, M. Tabata, S. Akashi, R and Sato, S. 2016. The complete chloroplast genome sequence of Zoysia matrella (L.) Merr. Crop Science 56: 1206-1212.
Wambugu, PW. Brozynska, M. Furtado, A. Waters, DL and Henry, RJ. 2015. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Scientific Reports 5: 13957.
Wang, R. Wang, X. Liu, K. Zhang, X.-J. Zhang, L.-Y and Fan, S-J. 2020. Comparative transcriptome analysis of halophyte Zoysia macrostachya in response to salinity stress. Plants 9: 458.
Yoo, S.-C. Oh, S-H and Park, J. 2021. Phylogenetic position of Daphne genkwa (Thymelaeaceae) inferred from complete chloroplast data. Korean Journal of Plant Taxonomy 51: 171-175.
Yu, TY. Yeam, DY. Kim, YJ and Kim, SJ. 1974. Morphological studies on Korean lawn grasses (Zoysia spp.). Journal of the Korean Society for Horticultural Science 15: 79-91.
Zerbino, DR and Birney, E. 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821-829.
Zhao, Q-Y. Wang, Y. Kong, Y-M. Luo, D. Li, X and Hao, P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: A comparative study. BMC Bioinformatics 12: S2.
|
|
|||||||||||||||||||||||||||||||||||||||