AbstractThe genus Chloranthus consists of 17 taxa with the two subgenera Chloranthus and Tricercandra. These two subgenera can be distinguished by the morphology of the androecium. In South Korea, C. japonicus and C. fortunei of the subgenus Tricercandra are distributed. Although C. fortunei is typically not branched, a variant that branches from the stem has recently been discovered. Here, we conducted morphological and molecular phylogenetic analyses to elucidate the interspecific relationships within the subgenus Tricercandra and to evaluate the taxonomic position of the variant of C. fortunei. Morphological analyses revealed distinct differences among C. angustifolius, C. japonicus, C. fortunei, and the variant of C. fortunei. However, distinguishing between C. holostegius and C. nervosus was challenging. A principal component analysis also showed that while C. angustifolius, C. japonicus, C. fortunei, and the corresponding variant clustered independently, C. holostegius and C. nervosus overlapped, rendering them indistinguishable. Molecular phylogenetic studies using nuclear ribosomal internal transcribed spacer (nrITS) regions and three plastid regions, rbcL, rpl20-rps12, and trnL-F, revealed that the subgenus Tricercandra was monophyletic. Within the subgenus Tricercandra, C. angustifolius formed a sister clade with C. fortunei in the combined three plastid gene tree, while C. japonicus displayed different phylogenetic relationships between the nrITS and plastid gene trees. In addition, neither C. holostegius nor C. nervosus was monophyletic, consistent with the morphological analyses. The variant of C. fortunei formed an independent subclade on the nrITS tree but was not distinguished from other C. fortunei on the plastid gene tree. Our study elucidated clear species delimitation among C. angustifolius, C. japonicus, and C. fortunei and suggested key morphological characters useful for distinguishing them. Furthermore, our findings highlighted the ambiguous species delimitation and phylogenetic uncertainty between C. holostegius and C. nervosus. While we demonstrated morphological differences between the typical C. fortunei and its variant, the limited molecular phylogenetic evidence implies a need for further study.
INTRODUCTIONThe genus Chloranthus, belonging to the family Chloranthaceae, is known to have approximately 17 taxa distributed across East and Southeast Asia (Wu, 1982). The family Chloranthaceae holds a significant position in the relationship between angiosperms. Chloranthaceae was previously recognized as a member of Piperales within the subclass Magnoliidae (Melchior, 1964; Takhtajan, 1969, 1980; Throne, 1974, 1992; Dahlgren, 1980; Cronquist, 1981, 1988; Okada, 1995). However, after the application of DNA sequencing in a phylogenetic study, the family Chloranthaceae was separated from Piperales, even from magnoliids, and is now considered an independent order, Chloranthales, in two recently released Angiosperm Phylogeny Group (APG) systems (Angiosperm Phylogeny Group, 2009, 2016). The order Chloranthales is regarded as one of the five major groups constituting Mesangiospermae, along with eudicots, monocots, magnoliids, and Ceratophyllales; thus, genomic data of Chloranthus species have been studied to reveal the evolutionary history of Mesangiospermae (Guo et al., 2021; Ma et al., 2021).
The infrageneric classification of the genus Chloranthus was proposed by Solms-Laubach (1869) based on growth forms into subshrubs (subgenus Fruticosi) and herbs (subg. Herbacei). Within the subg. Fruticosi, sections Triandri and Monandri were distinguished by the number of stamens, while within the subg. Herbacei, sections Brachyuri and Macronuri were distinguished by the elongation of the androecium (Table 1). However, Bentham and Hooker (1880) proposed a classification of the genus Chloranthus sensu lato into three sections, mainly focused on the morphology of the androecium rather than growth forms. They integrated sect. Triandri of subg. Fruticosi and sect. Brachyuri of subg. Herbacei (according to the Solms-Laubach classification) into the sect. Euchloranthus and treated sect. Monandri of subg. Fruticosi as sect. Sarcandra and sect. Macronuri of subg. Herbacei as sect. Tricercandra (Table 1). Conversely, Nakai (1930) proposed the three sections in Bentham and Hooker’s classification as three independent genera. More recently, Kong (2000a) divided the genus Chloranthus into two subgenera, Chloranthus and Tricercandra, based on the morphology of the androecium and established two sections, Chloranthus (sect. Triandri sensu Solms-Laubach) and Brachyuri (sect. Brachyuri sensu Solms-Laubach), in the subg. Chloranthus (Table 1).
The subg. Tricercandra sensu Kong, having an elongated tripartite androecium, consists of the six species C. japonicus, C. fortunei, C. angustifolius, C. nervosus, C. holostegius and C. coccineus, distributed only in East Asia. Chloranthus coccineus is a recently discovered species, having a morphology similar to that of C. fortunei but distinguishable by a longer peduncle and oval drupes, reddish leaf margins, and a crimson stamen connective apex (He et al., 2022). Except for recently discovered C. coccineus, many studies based on morphological, palynological, cytological, anatomical, and molecular phylogenetic evidence have been conducted on the genus Chloranthus, including the subg. Tricercandra (Swamy, 1953; Kuprianova, 1967; Endress, 1987; Todzia, 1993; Eklund, 1999; Kong, 2000a, 2000b, 2001; Kong and Chen, 2000; Kong et al., 2002; Zhang and Renner, 2003; Eklund et al., 2004; Zhang et al., 2011). The infrageneric classification of the genus Chloranthus based on the androecium morphology is supported by molecular phylogenetic evidence (Kong and Chen, 2000; Kong et al., 2002; Zhang and Renner, 2003; Zhang et al., 2011). Additionally, a variant discovered in island regions within South Korea exhibits a long tripartite androecium, and its flower and fruit are similar to those of C. fortunei of the subg. Tricercandra. However, this variant shows a branched stem, making it distinct from C. fortunei (Gray, 1859; Solms-Laubach, 1869; Nakai, 1930; Iwatsuki et al., 2006; Eom, 2007; Xia and Jeremie, 2007). Regarding the branching stem of C. fortunei, Kim (2007) noted that the stems of species in the genus Chloranthus are rarely branched. Eklund et al. (2004) also noted that some individuals of C. fortunei have two branches from the stem apex, but the discovered variants exhibit two branches from the stem apex as well as two branches from the main stem, possessing a total of five inflorescences (including the original one from the stem apex), representing a distinctive feature compared to previously reported variants of C. fortunei.
Here, we conducted a taxonomic and phylogenetic study of five species (excluding C. coccineus) of the subg. Tricercandra, along with the newly discovered variant of C. fortunei, based on morphological and molecular phylogenetic analyses. The specific goals of this study were to examine the species delimitation of the species within the subg. Tricercandra and to understand their phylogenetic relationships in an effort to determine whether the variant should be placed under C. fortunei or should be treated as an independent species.
MATERIALS AND METHODSTaxon samplingThe materials used in the experiment comprised a total of six taxa, including five taxa from the subgenus Tricercandra and a variant of C. fortunei (Online Supplementary Materials S1). These were fixed in the field using FAA or 70% ethanol, while some individuals were transplanted to a greenhouse at Kangwon National University. Voucher specimens were deposited into the Kangwon National University Herbarium (KWNU). Specimens from the following herbaria were examined: KWNU, HHU, CDBI, G, HUH, IBSC, JE, KEW, KUN, MBK, PE, PH, S, TAIF, and US (Appendix 1). Herbarium acronyms follow Index Herbariorum (Thiers, 2023).
Morphological analysesA morphological study was conducted based on collected herbarium specimens and/or their photos (Appendix 1), wherein 18 qualitative characters and 30 quantitative characters were examined. Due to the morphological characteristics of Chloranthus species, the vegetative organs, including the stem and leaf, still grow from the flowering time to the fruiting time, causing noticeable differences in certain quantitative characters. Hence, only the quantitative characters of the vegetative organs during the fruiting time were examined. In total, 30 quantitative characters (10 at the flowering time and 20 at the fruiting time) were examined. The 30 quantitative characters were measured, and the measurement methods used here are presented in Table 2 and Fig. 1. Based on the 30 quantitative characters, a principal component analysis (PCA) was conducted using PC-ORD (version 6) (McCune and Mefford, 2011) for both the flowering and fruiting times. Among the results, for principal components (PCs) 1, 2, and 3 with high contribution rates, the loading values of each character and the cumulative contribution rates were calculated. Scatter plots of individuals were generated in a two-dimensional space arrayed by PCs 1 and 2. To validate the significance of the quantitative characters extracted by the PCA analysis, the extracted quantitative characters were tested through an analysis of variance (ANOVA) and Bonferroni post-hoc test using SYSTAT (version 10, SPSS Inc., Chicago, IL, USA). Among the statistically significant characters, the differences among the five taxa were indicated by strings. For each character, taxa sharing the same string indicate no statistical difference, whereas those not sharing any string indicate statistical differences between them.
Molecular phylogenetic analysesWe selected 14 individuals from the five taxa, specifically four species of the subg. Tricercandra (excluding C. angustifolius) and the variant of C. fortunei. In addition to these 14 individuals, we downloaded previously reported sequences of 11 individuals, including the two subgenera Tricercandra and Chloranthus, from the NCBI GenBank, totaling 13 taxa and 26 individuals (Online Supplementary Materials S2). Sarcandra glabra was selected as an outgroup for all phylogenetic analyses. Genomic DNA was extracted using the DNeasy Plant Mini kit (Qiagen, Hilden, Germany). DNA amplification was conducted using a DNA thermal cycler (Biometra, Whatman Co., Göttingen, Germany). The nuclear ribosomal internal transcribed spacer (nrITS) region and three plastid markers, trnL-F, rpl20-rps12, and rbcL, were chosen, because those regions were used successfully for phylogenetic reconstruction in previous studies (Kong and Chen, 2000; Kong et al., 2002; Zhang and Renner, 2003; Zhang et al., 2011). The primers used for the polymerase chain reaction (PCR) amplification of nrITS, plastid trnL-F, rpl20- rps12, and rbcL, were obtained from previous studies, following their PCR profiles (White et al., 1990; Taberlet et al., 1991; Hasebe et al., 1994; Wen and Zimmer, 1996; Hamilton, 1999). The reaction solution used for amplification included 2.5 μL of 10× reaction buffer, 2.5 μL of 10 mM of dNTP mix, 0.5 μL each of 10 pmol forward and reverse primers, 0.2 μL of rTaq DNA polymerase (ELPIS Biotech Inc., Daejeon, Korea), 1 μL of template DNA, and 17.8 μL of distilled water, totaling 25 μl. The amplified PCR products were purified using a HiGene PCR purification kit (Solgent Co., Ltd., Seoul, Korea). Following purification, sequencing reaction processes were conducted using ABI PRISM BigDyeTM Terminator cycle sequencing kits (PE Applied Biosystems, Foster City, CA, USA), with the automatic DNA analyzer system ABI 3730xl DNA Analyzer (Applied Biosystems) utilized for a nucleotide sequencing analysis at Macrogene Inc. (Seoul, Korea). All sequences newly obtained in this study were deposited into the NCBI GenBank (Online Supplementary Materials S2). The obtained sequences were aligned using MAFFT (Katoh and Standley, 2013). Based on the aligned sequences, the maximum likelihood phylogenetic tree was constructed using RAxML (Stamatakis, 2006) with 1,000 bootstrap replicates and the substitution model GTRGAMMA.
RESULTSQualitative characters of the morphologyA total of 18 qualitative characters were observed for the six taxa, i.e., the five species of the subg. Tricercandra and the variant of C. fortunei (Table 3). When examining the morphological characters of the six taxa, it was observed that all taxa possessed fibrous roots. The inflorescence of all taxa was identified as a spike, and the color of the flowers was uniformly white across all taxa. The androecium was positioned on the outside of the apical part of the ovary in all taxa. The position of the ovary was identified as the inferior ovary position, and the shape of the seeds was either globose or ovoid, with all six taxa exhibiting similar morphologies in terms of these characters (Table 3). However, the six taxa were distinguished by branching, the shape of the cataphylls (in the stem), the leaf morphology, the shape of the bracteole (in the inflorescence), the anther positions, and the shapes of the ovary and fruit (Table 3).
Among the qualitative morphological characters, the key distinguishing characters varied across taxa. Chloranthus angustifolius have lanceolate to narrowly elliptic leaves, distinguishing them from other taxa with widely elliptic or obovate leaves. Chloranthus japonicus, which is widely distributed in East Asia, can be distinguished by the position of the anther on the flowers during the flowering time and by the shape of the bracteole on the inflorescence during the fruiting time (more obvious during the fruiting time). The anther of C. japonicus protrudes externally from the androecium, exhibiting a yellowish color around its white androecium, whereas in other taxa, especially in C. fortunei, the anther is encased within the androecium and is not readily apparent. Both C. nervosus and C. holostegius differed from other taxa within the subg. Tricercandra in having an ovate or oblong shape of the cataphyll of the stem, an opposite leaf arrangement that appears almost whorled, and a cuneate or obtuse leaf base. While a slight difference in the leaf margin was observed between these two taxa (serrate or dentate-serrate vs. coarsely serrate), distinguishing between them remains challenging. The typical C. fortunei and its variant can be distinguished from other taxa by their relatively long androecium and the 2-, 3-, or 4-toothed obovate shape of the bracteole on the inflorescence, which is ovate, semiorbicular, or triangular in other taxa, including C. angustifolius, C. japonicus, C. nervosus, and C. holostegius (Fig. 2A). Only branching of the stem revealed differences between the typical C. fortunei and its variant among the qualitative characters (Fig. 2B).
Principal component analyses based on quantitative charactersWe measured a total of 30 quantitative characters, distinguishing ten characters from the flowering time (codes 1 to 10) and twenty characters from the fruiting time (codes 11 to 30) (Table 2). The results are presented in Table 4. To examine the morphological variations between the taxa and verify the PC among the quantitative morphological characters, we conducted PCA based on the ten and twenty quantitative characters from the flowering time and fruiting time, respectively (Fig. 3). During the flowering time, the cumulative eigenvalues for PCs 1, 2, and 3, which had high loadings, were calculated and found to be 77.9% (Table 5). PC1, contributing 47.1%, showed a high correlation with the length of the inflorescence and peduncle (code 3), the length of the inflorescence (code 1), the number of inflorescences (code 6), and the length between flowers (code 5). PC2, with a 21.4% contribution, showed a correlation with the length of the androecium (code 7), the number of anthers (code 8), and the length of the ovary (code 9). PC3, contributing 9.3%, was correlated with the length of the peduncle (code 2), the number of flowers (code 4), and the width of the ovary (code 10). When PCs 1 and 2 were plotted in a two-dimensional space, the five taxa of the sub. Tricercandra and the variant of C. fortunei appeared to be distinguished; however, C. holostegius and C. nervosus overlapped slightly (Fig. 3A).
In the PCA result of the twenty quantitative characters at the fruiting time, the cumulative eigenvalues relative to the total covariance of PCs 1, 2, and 3 were found to be 63.9%, an outcome slightly lower than that at the flowering time (Table 6). PC1, accounting for 26.2% of the contribution, was correlated with the width of the leaf blade (code 19), the number of lateral veins (code 26), the length of the leaf blade (code 18), the number of teeth (code 24), and the length of the stem (code 11). PC2, with a contribution of 24.0%, was associated with the length of the petiole (code 21), the length and number of branches at the stem apex (codes 14 and 15, respectively), the number of leaves (code 25), and the number of branches at the central node (code 16). PC3, contributing 13.6%, was correlated with the number of nodes (code 12), the ratio of the leaf length to the width (code 20), the angle of the leaf apex (code 22), the length of the bracteole (code 28), and the number of bracteole lobes (code 27). In a two-dimensional plot of PCs 1 and 2, C. angustifolius, C. fortunei, and its variant were distinguished from other species. However, C. japonicus, C. holostegius, and C. nervosus overlapped in terms of the quantitative characters, making them indistinguishable regarding these characters at the fruiting time (Fig. 3B).
Statistical analyses of the extracted PCsWe conducted ANOVA on ten and fifteen characters of the flowering and fruiting times, extracted from the PCA results, finding that the number of anthers (code 8) was statistically insignificant, whereas the other 24 quantitative characters were confirmed as statistically significant (Online Supplementary Materials S3). The differences between the taxa for those 24 quantitative characters, excluding the number of anthers (code 8), are presented in Online Supplementary Materials S4. The five taxa of the subg. Tricercandra and the variant of C. fortunei were found to have statistically significant differences in five quantitative characters. The variant of C. fortunei differed statistically from the other five taxa in terms of the number and length of the branches at the stem apex (codes 14 and 15), the number of branches at the central node (code 16), the length of the petiole (code 21), and the number of lateral veins (code 26) (p < 0.05) (Fig. 4, Online Supplementary Materials S4).
In addition to the five characters that differed from all taxa, the variant of C. fortunei differed from the typical C. fortunei in terms of eight quantitative characters, in this case the length of the inflorescence (code 2), the length of the inflorescence and peduncle (code 3), the length between flowers (code 5), the number of inflorescences (code 6), the width of the leaf blade (code 19), the ratio of the leaf length to the width (code 20), the number of teeth (code 24), and the number of leaves (code 25) (Fig. 4). A total of ten quantitative characters, specifically the length of the peduncle (code 2), the length of the inflorescence and peduncle (code 3), the length between flowers (code 5), the number of inflorescences (code 6), the length of the androecium (code 7), the length of the ovary (code 9), the width of the ovary (code 10), the number of leaves (code 25), the number of bracteole lobes (code 27), and the length of the bracteole (code 28), showed statistically significant differences between C. japonicus and the variant of C. fortunei. Between C. japonicus and the typical C. fortunei, nine significantly different characters were found, specifically the length of the androecium (code 7), the length of the ovary (code 9), the width of the ovary (code 10), the number of nodes (code 12), the width of the leaf blade (code 19), the length of the petiole (code 21), the number of lateral veins (code 26), the number of bracteole lobes (code 27), and the length of bracteole (code 28). Only two characters, in this case the length of the peduncle (code 2) and the number of teeth (code 24), showed significant differences between C. holostegius and C. nervosus (Online Supplementary Materials S4).
Molecular phylogenetic analysesWe sequenced a total of four regions, consisting of nrITS and the three plastid regions rbcL, rpl20-rps12, and trnL-F to understand the phylogenetic relationships of the genus Chloranthus. The aligned length of nrITS was 676 bp, made up of the ITS1, 5.8S, and ITS2 regions. Among the three plastid regions, the aligned length of the rbcL gene was longest at 1,233 bp, followed by the trnL-F region (1,048 bp) and the rpl20-rps12 region (847 bp). The three plastid regions were concatenated into a single matrix, and the concatenated matrix was used for phylogenetic reconstruction. The results for both the nrITS and combined plastid gene trees indicated that the genus Chloranthus and the subg. Tricercandra were monophyletic (Fig. 5). However, the monophyly of the subg. Chloranthus collapsed in the plastid gene tree (Fig. 5B). Within the subg. Chloranthus, sister relationships between C. erectus and C. spicatus, between C. serratus and C. henryi, and between C. sessilifolius and C. oldhamii were recovered in both the nrITS and plastid gene trees, but the clade of C. sessilifolius, C. oldhamii, C. serratus, and C. henryi was a sister to the clade of the subg. Tricercandra, resulting in the collapse of the monophyly of the subg. Chloranthus in the plastid gene tree (Fig. 5B).
Within the subg. Tricercandra, monophyly of the subgenus was supported in both the nrITS and plastid gene trees with 84% and 53% support values, respectively (Fig. 5). Chloranthus nervosus and C. holostegius were not clearly distinguished and were clustered together. Among four individuals of C. nervosus, one individual formed a subclade with one C. holostegius (HUH168) in the nrITS tree (Fig. 5A), and it was sister to the clade of C. nervosus, C. holostegius, and C. japonicus in the plastid tree (Fig. 5B). These results imply an uncertain species delimitation between C. nervosus and C. holostegius. Among four individuals of C. japonicus, one individual (KWNU88286) was missing in the nrITS tree. In the nrITS tree, three individuals of C. japonicus (KWNU91691, KWNU56701, and one previously reported) formed a subclade, which was further a sister to the clade of C. fortunei and C. angustifolius (Fig. 5A), whereas the clade of C. japonicus, including three individuals newly sequenced in this study (KWNU91691, KWNU88286, and KWNU56701), was a sister to the clade of C. nervosus and C. holostegius in the plastid tree (Fig. 5B). These relationships were relatively poorly supported (66% in the nrITS tree and less than 50% in the plastid tree).
In both the nrITS and plastid trees, the close relationship between C. fortunei and C. angustifolius was strongly supported with 85% and 100% branch support values, respectively, but the topologies differed between the two trees. While C. angustifolius was a sister to all of the C. fortunei individuals, including the typical C. fortunei and its variants, in the plastid tree, it was a sister to the clade of three typical C. fortunei (KWNU76900, KWNU74253, and KWNU91770) in the nrITS tree (Fig. 5). Four individuals of the variant formed a subclade and were distinguished from other individuals of C. fortunei in the nrITS tree (Fig. 5A); however, they showed polytomy with other individuals of C. fortunei in the plastid tree (Fig. 5B). Both the nrITS and plastid trees consistently showed that the variants of C. fortunei were not clearly distinct from the typical C. fortunei, and the phylogenetic relationships between the typical C. fortunei and its variants remain unclear.
DISCUSSIONSpecies delimitation among the species of the subg. Tricercandra and the variant of C. fortuneiThrough our morphological analyses, it was demonstrated that C. angustifolius differed from other taxa based on the leaf morphology, specifically according to the lanceolate or narrowly elliptic leaves, leaf size, leaf apex, the number of lateral veins, and based on a character of the stem, in this case the number of nodes (Tables 3, 4). These differences were supported by the PCA and ANOVA results (Figs. 3 and 4, Online Supplementary Materials S4), enabling the identification of the species. However, it has been controversial as to whether C. holostegius and C. nervosus are independent species or should be treated as one species (Verdcourt, 1986, 1992; Kong, 2000a; Kong et al., 2002). In our study, they exhibited very similar morphologies. The differences between the two species were the leaf margin, the length of the inflorescence, and the number of teeth on the leaf margin (Tables 3, 4), which showed statistically significant differences in the ANOVA (Fig. 4, Online Supplementary Materials S4). The PCA results based on quantitative characters allowed for a slightly high resolution during the flowering time, albeit with some overlap; however, during the fruiting time, the characters overlapped entirely (Fig. 3), making them indistinguishable.
The close relationship between C. japonicus and C. fortunei has been reported in previous studies. Previous cytological studies suggested that C. fortunei (2n = 60) may have originated from an autotetraploid of C. japonicus (2n = 30) or from an allotetraploid between C. japonicus and the other species, such as C. serratus (Shinagawa and Tanaka, 1964; Hizume and Tanaka, 1982; Eom, 2007). However, our morphological analyses suggest a distant relationship between the two species, which was also supported by the PCA results. The most distinct differences during the flowering time, in this case the length of the androecium and the position and number of anthers, were observed here; C. japonicus had a significantly shorter androecium length averaging 4.8 mm, approximately half of that of C. fortunei (9.9 mm), and while C. japonicus had a total of two extrorse anthers located on the lateral stamens, C. fortunei had a total of four introrse anthers (Table 4). It has been reported that the shape of the bracteole can be used to distinguish between C. japonicus and C. fortunei during the fruiting time (Eom, 2007; Kim, 2007). Our study also confirmed that the shape of the bracteole, either triangular or semiorbicular cataphylls of the stem in C. japonicus and with a 2-, 3-, or 4-toothed obovate bracteole in C. fortunei, can be used for identification between the two species, and other morphological characters such as the length of the leaf sheath and the number of lateral veins observed during the fruiting period also allow for differentiation between the two taxa.
Chloranthus fortunei, distributed in South Korea and initially described as C. koreanus Nakai (1930), was later treated as C. fortunei var. koreanus (Nakai) Hiyama (1962). The taxonomic treatment was based on the characteristic of having slightly shorter central stamens in the tripartite androecium. However, Kim et al. (2000) argued for its treatment as C. fortunei, stating that the length of the central stamen shows variation and thus does not provide a clear distinction from C. fortunei. However, the variant of C. fortunei in this study that exhibits branching stems, not a shorter central stamen of C. fortunei var. koreanus, showed a difference in the qualitative characters compared to other taxa, including the typical C. fortunei. The results from quantitative traits showed that, apart from the trait of stem branching confirmed in the qualitative traits, the variant of C. fortunei differed from all other taxa in terms of the length of the petiole and the number of lateral veins, which are considered useful characters for distinguishing them during the fruiting time. Furthermore, the variant of C. fortunei differed from the typical C. fortunei in terms of certain floral characters, in this case the length of the peduncle and inflorescence, the length between flowers, and the number of inflorescences during the flowering time, and in terms of the vegetative characters of the width of the leaf blade, the number of teeth on the leaf margin, and the number of leaves during the fruiting time. Consequently, C. angustifolius, C. japonicus, and C. fortunei were clearly distinguished from other species based on both qualitative and quantitative characters, demonstrating clear species delimitation. However, the species delimitation between C. nervosus and C. holostegius remains ambiguous, although they can be distinguished from the other three species in the subg. Tricercandra. Additionally, the only qualitative character distinguishing the variant of C. fortunei from the typical C. fortunei was the branched stem, with statistically significant differences observed in several quantitative characters. Nonetheless, further observations in subsequent studies are deemed necessary to ascertain whether these characters are consistently maintained.
Phylogenetic relationships of the subg. Tricercandra and variant of C. fortuneiOur molecular phylogenetic analyses revealed that only the monophyly of the subg. Tricercandra was supported in both the nrITS and plastid gene trees (Fig. 5). However, C. angustifolius and C. fortunei showed a close relationship, consistent with previous studies (Kong and Chen, 2000; Kong et al., 2002; Zhang and Renner, 2003; Zhang et al., 2011). The sister relationship between the two species was confirmed even in the plastid genome-based phylogeny (Kang et al., 2022). Chloranthus japonicus formed a clade with C. holostegius and C. nervosus in the plastid gene phylogeny, in agreement with earlier studies (Kong and Chen, 2000; Kong et al., 2002; Zhang and Renner, 2003; Zhang et al., 2011). However, the nrITS phylogeny showed a sister relationship between C. japonicus and the clade of C. angustifolius and C. fortunei, a finding that differed from those in previous studies (Kong and Chen, 2000; Kong et al., 2002; Zhang and Renner, 2003; Zhang et al., 2011). The ambiguous species delimitation between C. holostegius and C. nervosus was also evident in molecular phylogenetic analyses. On the nrITS tree, the two species, represented by four individuals of C. nervosus and two of C. holostegius, failed to show monophyly. Notably, one of the four C. nervosus individuals was a sister to the entire subg. Tricercandra, indicating that C. nervosus was not monophyletic. Our morphological and molecular phylogenetic results support the previous opinion that C. holostegius should be treated as C. nervosus (Verdcourt, 1986, 1992; Kong, 2000a; Kong et al., 2002), but further research with a large number of individuals (most likely population level) and multiple genes (at least at the plastid genome level) is necessary to reveal the species delimitation and phylogenetic relationship between C. holostegius and C. nervosus.
Although the variants of C. fortunei, which exhibited differences in certain morphological characters from the typical C. fortunei, formed an independent subclade on the nrITS tree (Fig. 5A), they were not distinguished from the typical C. fortunei on the plastid gene tree, clustering together in a single clade (Fig. 5B). Notably, in the nrITS tree, three individuals of the typical C. fortunei newly sequenced in this study were separated from one individual of C. fortunei from China, forming a subclade with C. angustifolius. Intriguingly, the individual from China was a sister to the variants of C. fortunei. Moreover, five heterozygous nucleotides were found in the ITS2 region of the variant of C. fortunei, and the five nucleotides were ambiguously determined as R, Y, or W in the nrITS sequences. These heterozygous nucleotides may be evidence of hybridization, but finding only one individual of C. fortunei from China prohibits any inference with regard to the genetic variation of C. fortunei in China, making further speculation challenging.
In conclusion, our study suggests that the species delimitation of C. angustifolius, C. japonicus, and C. fortunei is relatively clear based on both qualitative and quantitative characteristics of the morphology, with statistically significant differences verified through PCA and ANOVA analyses. In contrast, the species delimitation of C. holostegius and C. nervosus remains uncertain, and the corresponding phylogenetic relationships were not resolved, suggesting that C. holostegius may not be an independent species distinct from C. nervosus. Cytonuclear discordance between the phylogenies based on nrITS and three plastid regions was observed within the genus Chloranthus, suggesting the need to provide a better definition of their phylogenetic relationships through further studies using genomic data. Moreover, the variant of C. fortunei, which uniquely possesses a branching stem distinct from the typical C. fortunei, has been identified with differences in several morphological characters. However, it is essential to observe whether these characters are consistently maintained. Given the limitations of a phylogenetic analysis due to insufficient sequence data from universal markers, there is a pressing need for validation through the use of genomic data in future studies.
ONLINE SUPPLEMENTARY MATERIALSS1–S4 are available at http://doi.org/10/11110/kjpt.2023.53.4.243.
S1. Specimens of the five taxa in the subg. Tricercandra and the variant of C. fortunei. A: C. angustifolius (HUH3403A), B: C. nervosus (MBK0190077), C: C. holostegius (HUH168), D: C. japonicus (TAIF257638), E: C. fortunei (HUH26039), F: variant of C. fortunei (KWNU91906)
kjpt-53-4-243-Supplementary-Material-S1.tif
S2. GenBank accession numbers and corresponding voucher specimens used in the phylogenetic analyses
S3. Results of analyses of variance (ANOVA) using 25 principal components. Each row is a separate ANOVA of a single morphological character.
S4. Significant quantitative characters extracted by a principal component analysis (except for character 8)
kjpt-53-4-243-Supplementary-Material-S2-S4.xlsx
Fig. 1.Measurement of the quantitative characters of the subg. Tricercandra and variant of Chloranthus fortunei. A. Habit. B. Leaf. C. Inflorescence. D. Androecium. E. Fruit. F. Bracteole. 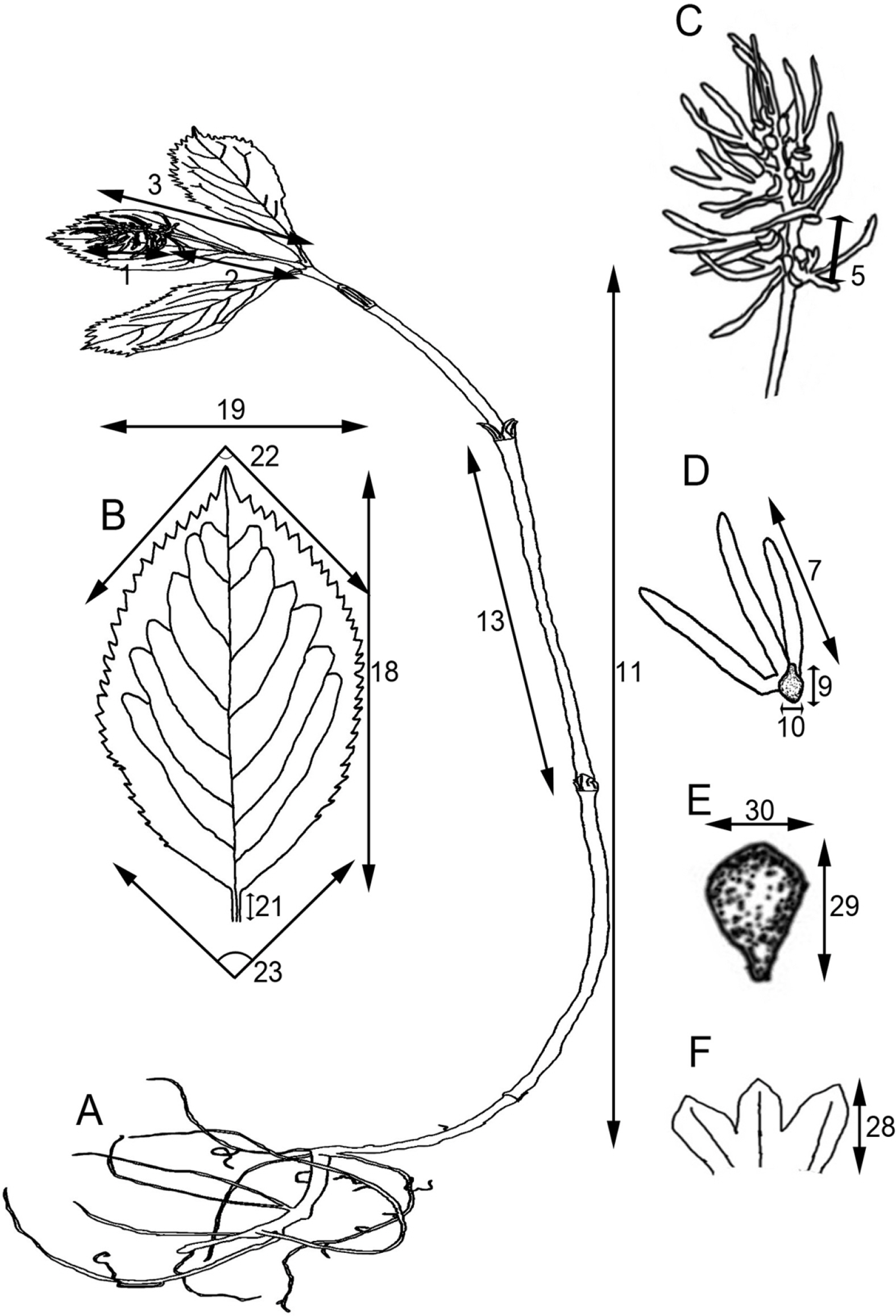
Fig. 2.Different morphological traits on the bracteole and stem of the subg. Tricercandra and variant of Chloranthus fortunei. A. Shape of the bracteole. B. Type of stem. 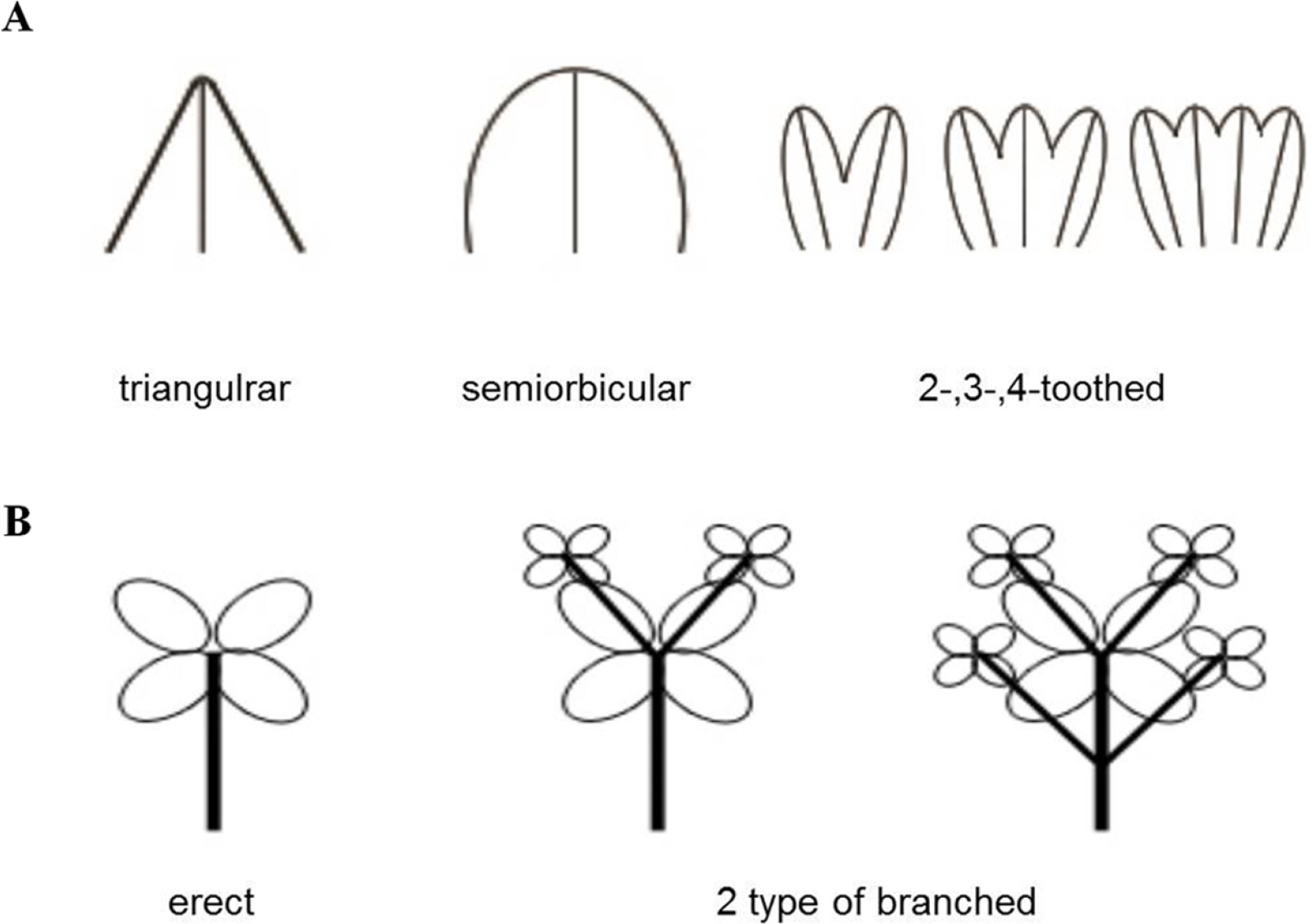
Fig. 3.Plots of principal components 1 and 2 based on quantitative morphological characters of the subg. Tricercandra and variant of Chloranthus fortunei. A. Principal component analysis (PCA) results of 48 individuals based on quantitative characters at the flowering time. B. PCA results of 71 individuals based on quantitative characters at the fruiting time. a, C. angustifolius; f, C. fortunei; h, C. holostegius; j, C. japonicus; n, C. nervosus; s, variant of C. fortunei. 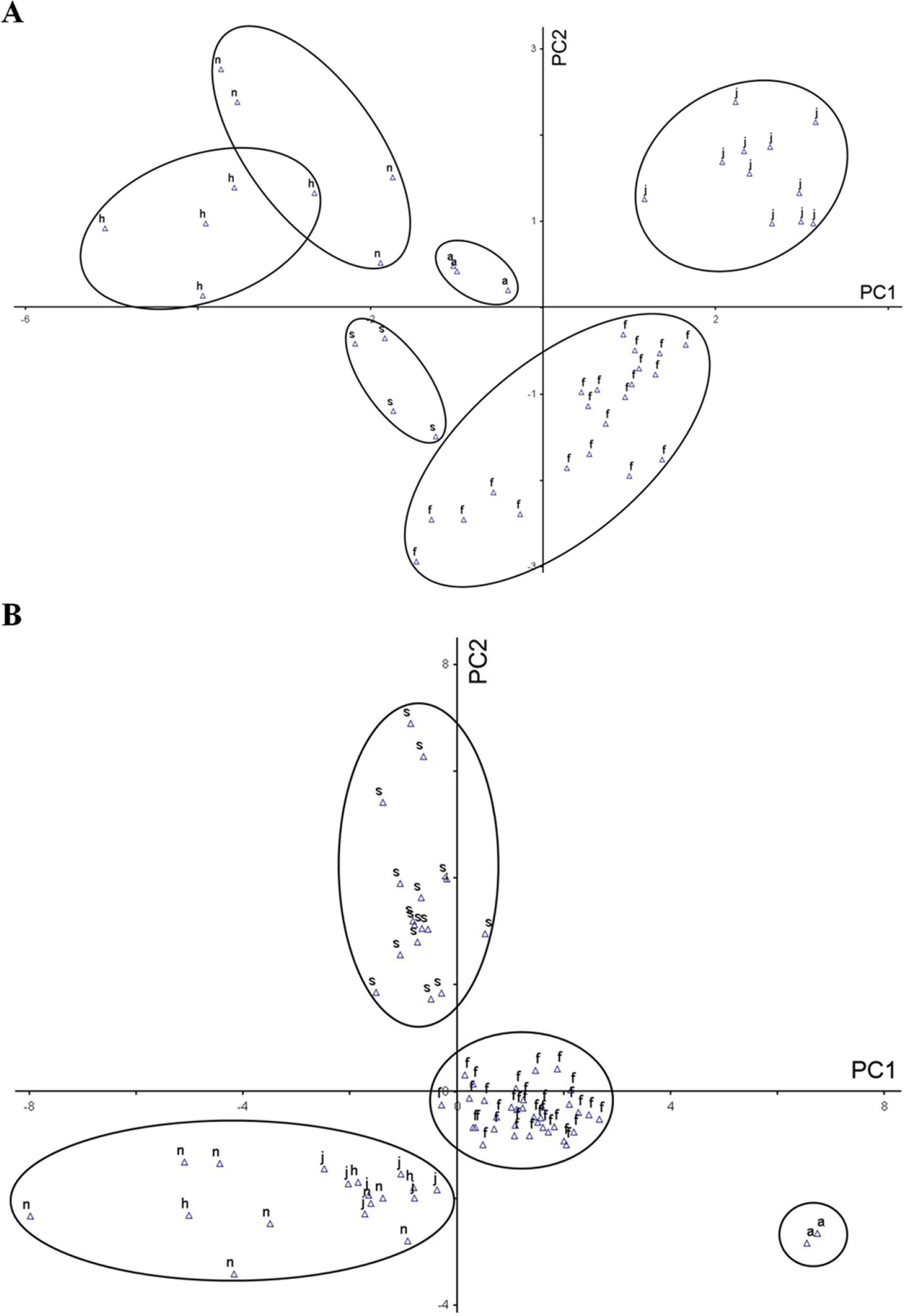
Fig. 4.Box plots of six quantitative characters showing differences among different taxa. Codes 3, 5, and 7 represent characters at the flowering time, while codes 20, 21, and 28 represent characters at the fruiting time. Significance levels were analyzed by ANOVA and Bonferroni post hoc tests. All six characters were significant (P-value < 0.001). Regarding the letters at the top of the box plot, identical letters shared between two taxa indicate no statistical difference between them. a, Chloranthus angustifolius; f, C. fortunei; h, C. holostegius; j, C. japonicus; n, C. nervosus; s, variant of C. fortunei. 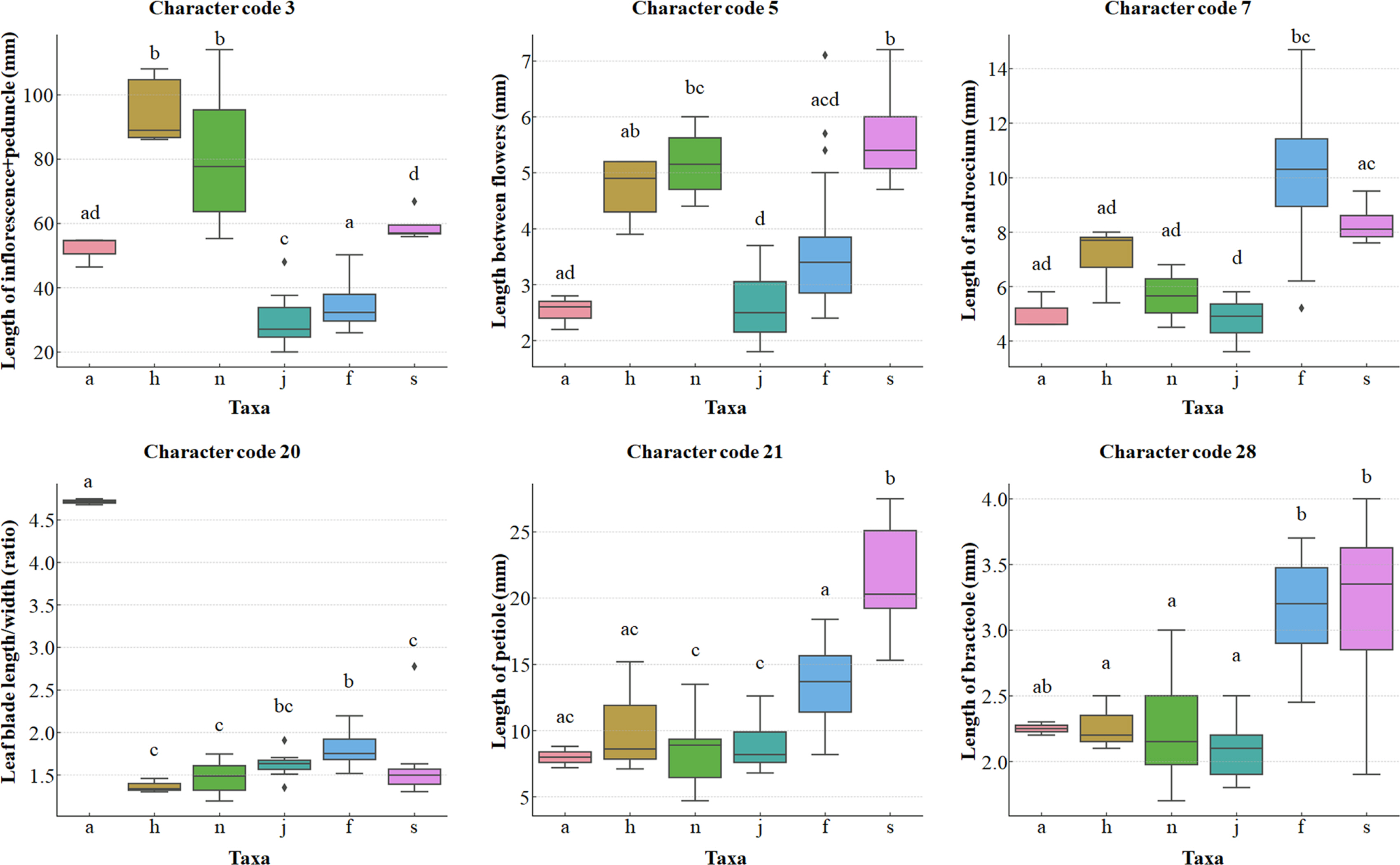
Fig. 5.Maximum likelihood trees of the genus Chloranthus based on sequences of the nuclear ribosomal internal transcribed spacer (nrITS) and three plastid regions. Phylogenetic reconstruction was conducted using RAxML. Bootstrap values greater than 50% are indicated near branches. A. Phylogenetic tree based on nrITS sequences. B. Phylogenetic tree based on concatenated sequences of the plastid rbcL, rpl20- rps12, and trnL-F regions. 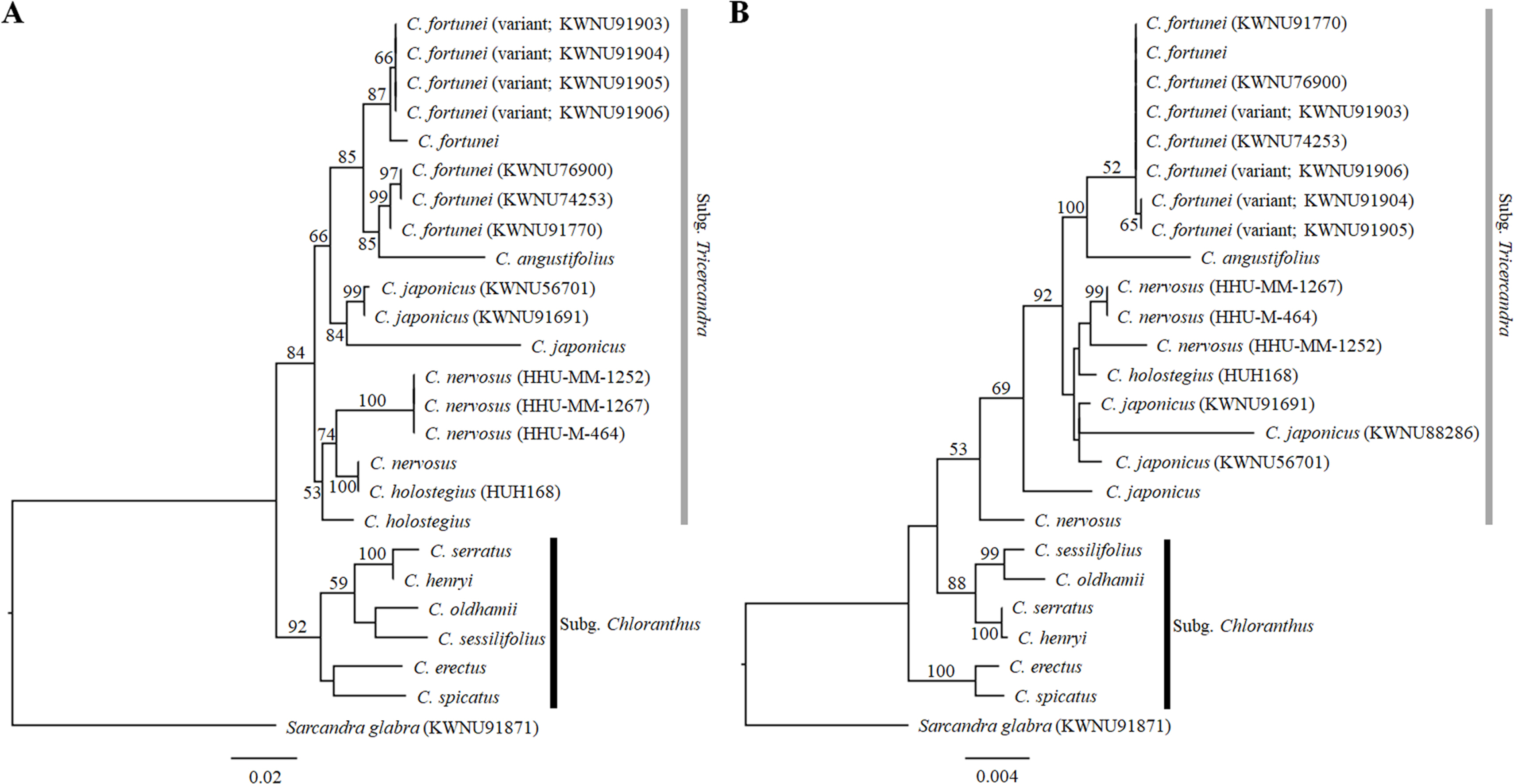
Table 1.Comparison of four infrageneric classifications of the genus Chloranthus sensu lato.
Table 2.Quantitative characters of the subg. Tricercandra and variant of Chloranthus fortunei measured in this study. Table 3.Qualitative characters of the subg. Tricercandra and variant of Chloranthus fortunei. Table 4.Quantitative characters of the subg. Tricercandra and variant of Chloranthus fortunei. Table 5.Loading values of the three principal components using 10 quantitative characters of the subg. Tricercandra and variant of Chloranthus fortunei at the flowering time. Table 6.Loading values of the three principal components using 20 quantitative characters of the subg. Tricercandra and variant of Chloranthus fortunei at the fruiting time LITERATURE CITEDAngiosperm Phylogeny Group. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society 161: 105-121.
Angiosperm Phylogeny Group. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181: 1-20.
Bentham, G and Hooker, JD. 1880. Genera Plantarum. 3: Reeve & Co., London. 133-135.
Cronquist, A. 1981. An Integrated System of Classification of Flowering Plants. Columbia University Press, New York. 1262 pp.
Cronquist, A. 1988. The Evolution and Classification of Flowering Plants. 2nd ed. The New York Botanical Garden, New York. 555 pp.
Dahlgren, R. M. T. 1980. A revised system of classification of the angiosperms. Botanical Journal of the Linnean Society 80: 91-124.
Eklund, H. 1999. Big survivors with small flowers: fossil history and evolution of Laurales and Chloranthaceae. PhD dissertation,. Uppsala University, Uppsala, Sweden. 52 pp.
Eklund, H. Doyle, JA and Herendeen, PS. 2004. Morphological phylogenetic analysis of living and fossil Chloranthaceae. International Journal of Plant Sciences 165: 107-151.
Endress, P.K. 1987. The Chloranthaceae: reproductive structures and phylogenetic position. Botanische Jahrbücher Systematik 109: 153-226.
Eom, J. A. 2007. A taxonomic study of the genus Chloranthus (Chloranthaceae) in Korea. MSc thesis,. Chungbuk National University; Cheongju, Korea. 38.
Gray, A. 1859.
Tricercandra & T. Fortunei
. Memoirs of American Academy of Arts and Sciences, New Series 6: 405.
Guo, X. Fang, D. Sahu, S. K. Yang, S. Guang, X. Folk, R. Smith, S. A. Chanderbali, A. S. Chen, S. Liu, M. Yang, T. Zhang, S. Liu, X. Xu, X. Soltis, P. S. Soltis, D. E and Liu, H. 2021.
Chloranthus genome provides insights into the early diversification of angiosperms. Nature Communications 12: 6930.
Hamilton, M.B. 1999. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Molecular Ecology 8: 521-523.
Hasebe, M. Omori, T. Nakazawa, M. Sano, T. Kato, M and Iwatsuki, K. 1994.
rbcL gene sequences provide evidence for the evolutionary lineages of leptosporangiate ferns. Proceedings of the National Academy of Sciences of the United States of America 91: 5730-5734.
He, L.-H. Zhao, X.-L. Yu, J and Zhang, Z. 2022.
Chloranthus coccineus (Chloranthaceae), a new species from Hubei, China. Phytotaxa 566: 279-289.
Hiyama, K. 1962. Miscellany from Makino herbarium (9). Jpn. J. Bot 37: 125-128.
Hizume, M and Tanaka, A. 1982. Karyomorphological studies on three Japanese species of Chloranthus
. La Chromosomo 25: 735-741.
Iwatsuki, K. Boufford, D. E and Ohba, H. 2006. Chloranthaceae. In Flora of Japan. IIa. Angiospermae Dicotyledoneae Archichlamydeae (a). Iwatsuki, K (ed.), Kodansha, Tokyo. 364-365.
Kang, J.-S. Kim, B.-Y and Yoo, K.-O. 2022. The complete plastid genome sequence of Chloranthus fortunei (A. Gray) Solms-Laub. in Chloranthaceae. Mitochondrial DNA Part B Resources 7: 1829-1833.
Katoh, K and Standley, D. M. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30: 772-780.
Kim, C. H. 2007. Chloranthaceae. The Genera of Vascular Plants of Korea. Park, C. W (ed.), Academy Publishing Co., Seoul. 148-149.
Kim, C. H. Kim, T. J and Sun, B.-Y. 2000. Taxonomic identities of some endemic Korean vascular plants. Korean Journal of Plant Taxonomy 30: 355-361.
Kong, H. Z. 2000a. Systematics of the genus Chloranthus Swartz (Chloranthaceae). PhD dissertation,. Institute of Botany, Chinese Academy of Sciences, Beijing. 116 pp.
Kong, H.-Z. 2000b. Karyotypes of Sarcandra Gardn. and Chloranthus Swartz (Chloranthaceae) from China. Botanical Journal of the Linnean Society 132: 327-342.
Kong, H.-Z. 2001. Comparative morphology of leaf epidermis in the Chloranthaceae. Botanical Journal of the Linnean Society 136: 279-294.
Kong, H.-Z and Chen, Z.-D. 2000. Phylogeny in Chloranthus Swartz (Chloranthaceae) inferred from sequence analysis of nrDNA ITS region. Acta Botanical Sinica 42: 762-764.
Kong, H.-Z. Chen, Z.-D and Lu, A.-M. 2002. Phylogeny of Chloranthus (Chloranthaceae) based on nuclear ribosomal ITS and plastid trnL-F sequence data. American Journal of Botany 89: 940-946.
Kuprianova, L.A. 1967. Palynological data for the history of the Chloranthaceae. Pollen et Spores 6: 95-100.
Ma, J. Sun, P. Wang, D. Wang, Z. Yang, J. Li, Y. Mu, W. Xu, R. Wu, Y. Dong, C. Shrestha, N. Liu, J and Yang, Y. 2021. The Chloranthus sessilifolius genome provides insight into early diversification of angiosperms. Nature Communications 12: 6929.
McCune, B and Mefford, M. J. 2011. PC-ORD. Multivariate Analysis of Ecological Data. Version 6. MjM Software, Clenden Beach, OR
Melchior, H. 1964. A Engler’s Syllabus der Pflanzenfamilien. Angiospermen. Piperales. 2/17. Borntraeger, Berlin. 147-151.
Nakai, T. 1930. Flora Sylvatica Koreana. Vo. 18. Government General of Chosen, Seoul. 20 pp.
Okada, H. 1995. Karyological studies of four genera of the Chloranthaceae. Plant Systematics and Evolution 195: 177-185.
Shinagawa, T and Tanaka, R. 1964. Chromosomes of Chloranthus fortunei (A. Gray) Solms. Journal of Japanese Botany 39: 145-148.
Solms-Laubach, H. A. 1869. Chloranthaceae. Prodromus Systematis Naturalis Regni Vegetabilis. 16: de Candolle, A (ed.), Victoris Masson et Filii, Paris. 474-477.
Stamatakis, A. 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688-2690.
Swamy, B. G. L. 1953. The morphology and relationships of the Chloranthaceae. Journal of the Arnold Arboretum 34: 375-411.
Taberlet, P. Gielly, L. Pautou, G and Bouvet, J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105-1109.
Takhtajan, A. 1969. Flowering Plants. Origin and Dispersal. Oliver & Boyd, Edinburgh. 310 (English edition translated by C. Jeffrey)..
Takhtajan, A.L. 1980. Outline of the classification of flowering plants (Magnoliophyta). Botanical Review 46: 225-359.
Thiers, B. 2023. Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. Retrieved Nov. 17, 2023, available from http://sweetgum.nybg.org/science/ih/..
Thorne, R. F. 1992. An updated phylogenetic classification of the flowering plants. Aliso 13: 365-389.
Todzia, C. A. 1993. Chloranthaceae. The Families and Genera of Vascular Plants. 2: Kubitzki, K. Rohwer, J. G and Brittrich, V. Springer, Berlin. 197-200.
Verdcourt, B. 1986. Chloranthaceae. Flora Malesiana. Series 1, Vol. 10, Part 2. van Steenis, C. G. G. J (ed.), Kluwer Academic Publishers, Dordrecht. 133-144.
Verdcourt, B. 1992. Chloranthaceae. Flora of Thailand. 5: Part 4.. Smitinand, T. Larsen, K (eds.), The Forest Herbarium, Royal Forest Department, Bangkok. 424-430.
Wen, J and Zimmer, E. A. 1996. Phylogeny and biogeography of Panax L. (the Ginseng genus, Araliaceae): Inferences from ITS sequences of nuclear ribosomal DNA. Molecular Phylogenetics and Evolution 6: 167-177.
White, T. J. Bruns, T. Lee, S and Taylor, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. Innis, M. A. Gelfand, D. H. Sninsky, J. J. White, T. J (eds.), Academic Press, San Diego, CA. 315-322.
Wu, K. F. 1982. Chloranthaceae. Flora Reipublicae Popularis Sinicae. 20: Tseng, Y. -C (ed.), Science Press, Beijing. 77-97.
Xia, N and Jeremie, J. 2007. Chloranthaceae. Flora of China. 4: Wu, Z. Y. Raven, P. H and Hong, D. Y. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis, MO. 132-137.
APPENDICESAppendix 1.
Information about the specimens subjected to the morphological analyses. Vouchers included the herbarium acronym and voucher number
Chloranthus angustifolius Oliver CDBI0010046, K0000802651, KUN0200084, HUH3403A, IBSC130783, PE00002946
Chloranthus fortunei (A. Gray) Solms HUH26039, KUN0200188, KUN0200190, KWNU91770, KWNU91771, KWNU91772, KWNU91773, KWNU91774, KWNU91775, KWNU91776, KWNU91777, KWNU91778, KWNU91779, KWNU91780, KWNU91781, KWNU91782, KWNU91783, KWNU91784, KWNU91785, KWNU91786, KWNU91787, KWNU91788, KWNU91789, KWNU91790, KWNU91791, KWNU91792, KWNU91793, KWNU91794, KWNU91795, KWNU91796, KWNU91797, KWNU91798, KWNU91799, KWNU91800, KWNU91801, KWNU91802, KWNU91803, KWNU91804, KWNU91805, KWNU91806, KWNU91807, KWNU91808, KWNU91809, KWNU91810, KWNU91811, KWNU91812, KWNU91813, KWNU91814, KWNU91815, KWNU91816, KWNU91816, KWNU91817, KWNU91818, KWNU91819, KWNU91820, KWNU91821, KWNU91822, KWNU91823, S12-27258
Variant of C. fortunei KWNU91872, KWNU91873, KWNU91874, KWNU91875, KWNU91876, KWNU91877, KWNU91878, KWNU91879, KWNU91880, KWNU91881, KWNU91882, KWNU91883, KWNU91884, KWNU91885, KWNU91886, KWNU91887, KWNU91888, KWNU91889, KWNU91890, KWNU91891, KWNU91892, KWNU91893, KWNU91894, KWNU91895, KWNU91896, KWNU91897, KWNU91898, KWNU91899, KWNU91900, KWNU91901, KWNU91902, KWNU91903, KWNU91904, KWNU91905, KWNU91906, KWNU91907, KWNU91908, KWNU91909
Chloranthus holostegius (Handel-Mazzetti) Pei & Shan CDBI0010088, CDBI0010089, CDBI0010090, CDBI0010095, HUH168, IBSC0130996, IBSC0131003, IBSC0131004, KUN0840091
Chloranthus japonicus Sieb. HUH1339, HUH5936B, HUH22609, HUH26389, KWNU05500, KWNU05524, KWNU56701, KWNU61894, KWNU61896, KWNU67263, KWNU70473, KWNU71482, KWNU71540, KWNU72382, KWNU72708, KWNU76553, KWNU85338, KWNU85585, TAIF257638
Chloranthus nervosus Coll. et Hemsl. HHU-M-464, HHU-MM-1252, HHU-MM-1267, HUH838, HUH90-708, HUH96-773, HUH97-696, KUN0975628, MBK0107842, MBK0190077
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||