Meiotic chromosome numbers of five Carex taxa in Korea (Cyperaceae)
Article information
Abstract
Carex L. (Cyperaceae) is the largest angiosperm genus in the temperate zones with more than 2,000 species worldwide. Unusual chromosome structures, called holocentric chromosomes, have been postulated to contribute to species diversity in the genus. In Korea, this genus has the greatest number of species, but chromosome information as it pertains to the taxa is mostly unknown. Here, we report meiotic chromosome numbers of five Carex taxa in Korea. The following observations are made: Carex jaluensis Kom. (n = 27II, 28II, 29II, 30II), C. japonica Thunb. (n = 28II, 29II), C. planiculmis Kom. (n = 30II), C. miyabei Franch. (n = 33II, 36II), C. neurocarpa Maxim. (n = 51II, 53II, 54II). Except for C. planiculmis, all of the species exhibit variations in chromosome numbers within individuals and/or taxa. The findings with regard to chromosome number diversity in Carex suggest that chromosome number variation (aneuploidy, agmatoploidy and/or symploidy) plays an important role in the richness of the species in the genus. Further cytological investigations are needed for a better understanding of sedge diversity in Korean flora.
Genus Carex L. (Cyperaceae), characterized with unisexual flowers and saclike structures in pistillate flowers (perigynia), is the largest angiosperm genus in the temperate zone with more than 2,000 species worldwide (Reznicek, 1990; Global Carex Group, 2015). The genus occurs in broad range of habitats from high mountains to riversides, and from sunny to shady areas (Egorova, 1999; Global Carex Group, 2015). One of the most intriguing characteristics, along high species diversity in the genus, is extremely broad variations in chromosome numbers ranging from n = 6 to n = 66, with every haploid number between n = 6 and n = 48 (Tanaka, 1949; Davies, 1956; Roalson, 2008; Hipp et al., 2009). The cytogenetic variance has been postulated to be associated with non-localized centromeres, holocentric chromosomes (Luceño and Guerra, 1996; Hipp et al., 2010). Unlike monocentric chromosomes, holocentric chromosomes can be fragmented and fused during cell divisions, which facilitates chromosome number increases (agmatoploidy) and/or decreases (symploidy) (Luceño and Guerra, 1996; Hipp et al., 2013). Because chromosome number variation in Carex is not necessarily associated with DNA duplication and/or deletion, it is called agmatoploidy or symploidy distinguished from aneuploidy (Nishikawa et al., 1984; Luceño and Guerra, 1996; Chung et al., 2011). Chromosome investigations have provided important information on taxonomy and phylogeny of Carex (Rothrock et al., 2009; Yano et al., 2010), and high variations in chromosome numbers have been hypothesized to play an important role on the rapid speciation in recently diverged lineages in the genus (Hipp, 2007; Hipp et al., 2009, 2010; Chung et al., 2012; Escudero et al., 2012).
In Korea, Carex is the most species-rich genus with about 180 native taxa, and Cyperaceae (sedge family) is the second largest family after Asteraceae (sunflower family) (Kim et al., 2008; Park et al., 2016). Although chromosome number information is critical to understand Carex diversity, chromosome investigations on Korean Carex taxa have not been actively conducted. Among about 180 Korean native Carex, only 18 taxa have chromosome numbers reported (Kim, 2006; Lee and Kim, 2008; Chung et al., 2013; Chung et al., 2016, 2017; Chung et al., 2018). The studies demonstrate polyploidy in Carex section Siderostictae Franch. ex Ohwi and continuous number variations within and/or among individuals or species in most of the sections investigated. In addition, a basal linage in Carex, sect. Siderosticatae, exhibits large chromosomes (about 2–4 μm long) with small numbers (2n = 12), whereas recently diverged linages have smaller chromosomes (less than 1 μm long) with high numbers and continuous number variations (Chung et al., 2013; Yano et al., 2014; Jiménez-Mejías et al., 2016; Chung et al., 2017).
In this paper, we report meiotic chromosome numbers of five Carex species from Korean populations and discuss their taxonomic, cytological significances.
Materials and Methods
The chromosome numbers of five Carex species from Korean populations were analyzed. To determine meiotic chromosome numbers, young spikes were fixed in natural habitats in May and June in 2015, 2016, and 2018. The methods for chromosome observation mainly followed Rothrock and Reznicek (1996) and Chung et al. (2016) using a fixing mixture of methanol, chloroform, and propionic acid (6:3:2). Fixed anthers were quashed in 1% or 2% acetic-orcein and observed at 1,000× magnification (Nikon Eclipse 50i, Nikon, Tokyo, Japan). To have acute analyses, each meiotic chromosome figure was drawn and photographed. In addition, meiotic chromosome numbers (n) were determined after observations of at least three pollen mother cells per sample. All voucher specimens were collected in June and July 2018 and stored at the Korea National Arboretum Herbarium (KH) (Table 1).
Results and Discussion
Meiotic chromosome numbers studied are tabulated with previous records (Table 1), and representative meiotic chromosome figures are presented (Fig. 1). All chromosomes are less than 1 μm long, none of the species exhibit distinct primary constriction, and only bivalents are observed.
-
Carex jaluensis Kom. (n = 27II, 28II, 29II, 30II) (Fig. 1A–C) – Sect. Anomalae J. Carey
This is the first report of chromosome number for C. jaluensis, n = 27II, 28II, 29II, 30II. The chromosome number variation within an individual and/or species is not unusual in Carex (Chung et al., 2011). There is always some room for technical, artificial mistakes, but it is also possible to see more than one chromosome number per taxon in holocentric chromosome processing organisms (Hoshino, 1981; Luceño and Guerra., 1996; Chung et al., 2011). The species occurs in Korea, China, and Russia; and in Korea it is found mainly in the Middle East regions (Park et al., 2016). The chromosome number variation within species is more closely related with morphological diversity and diverging time (genetic diversity) than geographic diversity (Hipp et al., 2010; Chung et al., 2012). Further chromosome investigations with geographic and morphological features are required for the species. Most species in the sect. Anomalae occur in Australia (Dai et al., 2010).
-
C. japonica Thunb. (n = 28II, 29II) (Fig. 1D) – Sect. Molliculae Ohwi
In an individual of C. japonica, two meiotic chromosome numbers n = 28II, 29II are observed, which is the first count from a Korean population. Previous chromosome counts for the species are made with Japanese individuals, 2n = 62 (Table 1) (Hoshino et al., 2011). The species is commonly found in forests and sunny grasslands in China, Japan and Korea (Dai et al., 2010; Hoshino et al., 2011; Park et al., 2016). Although the species distributes widely, variation in morphological characters is not significant. Various chromosome numbers for the species might be related with broad geographic distribution or recent speciation events (Chung et al., 2012). To understand the chromosome variation in the species, taxonomic and phylogenetic studies besides cytological research with more population samples covering all the distribution areas should be conducted.
-
C. planiculmis Kom. (n = 30II) – Sect. Molliculae
We observed meiotic chromosomes of n = 30II from C.planiculmis, which is incongruent with previous reports, 2n = 62 (Hoshino et al., 2011). The species is only found in East Asia (North East China, Japan, Korea, Far East Russia), growing in wet places in the forest (Dai et al., 2010; Hoshino et al., 2011; Park et al., 2016). Because only two chromosome number counts have been made for the species sampled from only the two geographic regions, additional investigations should be conducted to determine chromosome number variation range in the species. In the section Molliculae, Hoshino et al. (2011) reported chromosome numbers for C. doniana Spreng. (2n = 62), C. planiculmis (2n = 62), C. japonica (2n = 62), and C. mollicula Boott (2n = 56, 66, 68, 70). About 20 species occur in East and South East Asia with high diversity in China, in which 18 Carex sect. Molliculae species occur including nine endemics (Dai et al., 2010).
-
C. miyabei Franch. (n = 33II, 36II) – Sect. Carex
The meiotic chromosome number for the species is n = 33II and 36II. This is the second count from Korean populations and incongruent from the both previous reports from a Japanese population, 2n = 90 (Tanaka, 1948) and Korean populations, 2n = 84 (Chung et al., 2018). The species has been considered endemic to Japan until the very recently (Hoshino et al., 2011). However, the species occurs throughout Korea and has been often misidentified as C. glabrescens (Kük) Ohwi (Im et al., 2008; Park et al., 2016). Variations in morphological characters such as perigynium shapes and surface features have made it hard to distinguish the two taxa from each other. Chromosome numbers of C. glabrescens are unknown. Chromosome number variation in the section Carex has been found (ex. C. drymophila Turcz. ex Steud.) (Chung et al., 2018). Taxonomic and cytological research should be conducted to understand the two morphologically confusing taxa in the section.
-
C. neurocarpa Maxim. (n = 51II, 53II, 54II) (Fig. 1E, F) - Sect. Phleoideae Meinsh.
The meiotic chromosomes of C. neurocarpa observed vary, n = 51II, 53II 54II. Only one number is congruent from the previous reports from Japanese and Korean populations as n = 54II (Tanaka, 1937; Chung et al., 2018). Chromosome numbers of n = 51II and 53II are first counts for the species. C. neurocarpa grows mainly in wet places along ponds and riversides in China, Japan, Korea, and Far East Russia (Park et al., 2016). Although the species occurs widely in East Asia, by well-developed wings in perigynia, the species is distinguished from the other taxa in the section (Dai et al., 2010; Hoshino et al., 2011; Park et al., 2016). Section Phleoideae occurs in Asia and consists of about nine taxa, and the monophyly of the section is tested although taxon sampling is incomplete (Dai et al., 2010; Hoshino et al., 2011; Jiménez-Mejías et al., 2016). Among the nine taxa in the section, chromosome numbers of six taxa have been reported with variations in three taxa (Dai et al., 2010; Hoshino et al., 2011). Although the section is small only with nine taxa, variations in chromosome numbers and major morphological characters exhibit. The character variations make the section a good model group to test cytological, morphological, and geographic character evolution in Asian Carex group. In particular, analyzing chromosome number variations of Phleoideae in a phylogenetic framework will provide critical information to understand cytological evolution in Asian, bisexual spike (androgynous) Carex groups.
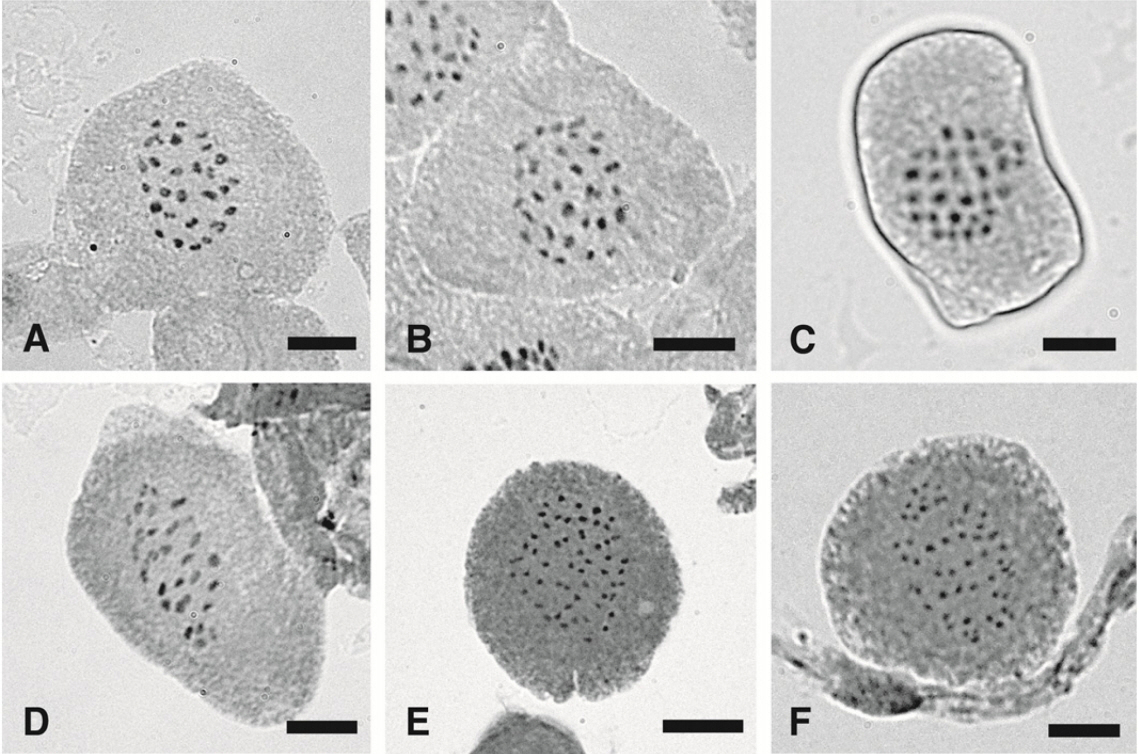
Photomicrographs of Carex meiotic metaphase chromosomes. A. Carex jaluensis (n = 28II, Chung 1129). B. C. jaluensis (n = 29II, Chung 1129). C. C. jaluensis (n = 30II, Chung 1134). D. C. japonica (n = 29II, Chung 1604). E. C. neurocarpa (n = 51II, Chung 5143). F. C. neurocarpa (n = 53II, Chung 5143). Scale bars = 5 μm.
All the species, except C. planiculmis, exhibit variations in chromosome numbers within individuals and/or taxa. The results of chromosome number diversity in Carex suggest that chromosome number variation (aneuploidy, agmatoploidy, and/or symploidy) might have played an important role in species richness in the genus. The investigations on chromosome characters of the species in a phylogenetic phylogenetic framework will hypothesize speciation mechanisms. To understand Carex species diversity, long-term and continuous efforts should be made to investigate chromosome information of Carex in spite of difficulty on conducting research with only living plant materials.
Acknowledgements
We thank Tomomi Masaki (Okayama University of Science) for suggestions on chromosome observations and two anonymous reviewers for critical comments. This study was supported in part by the National Research Foundation of Korea (NRF-2018R1A2B6008851).
Notes
Conflict of Interest
The authors declare that there are no conflicts of interests.