Ulleungdo Island is an oceanic island located in the East Sea, 130 km east of the eastern coast of the Korean peninsula and 280 km northwest from the western coast of Japan. The island is very small, with a size of about 73 km2. The island originated from the seafloor as a result of volcanic activity and emerged above sea level about 1.8 million years ago (Kim, 1985). Since the island was formed, it has been isolated from the neighboring mainland even during periods of glaciation (Kim et al., 2000). Seonginbong Peak (elevation 984 m), the island’s highest peak, is located at the center of the island and comprises five major ridges with very steep slopes. With the influence of the maritime environment, broadleaf evergreen plants occupy the lowland of the island, whereas broadleaf deciduous forests are well developed in mountainous areas above 300 m. A recent survey of island flora shows that there are 484 native vascular plants belonging to 116 families, 40 of which are endemic to the island (Sun et al., 2014).
The number of endemic taxa on Ulleungdo Island is very high given its small area. Each of the endemic taxa is known to have evolved via anagenetic speciation, in which a progenitor population dispersed from the neighboring mainland and evolved into a new species (Sun and Stuessy, 1998). Anagenetic speciation presumes a single introduction of the ancestral population to the island. Isolation plays an important role in the process, as frequent immigration and establishment of the progenitor species would prevent an insular population from splitting into a separate lineage. Pfosser et al. (2002), in a study of two Acer species endemic to Ulleungdo, described two contrasting cases: A. takesimense Nakai, which descended from a single introduction, and A. okamotoanum Nakai, which was derived from progenitors that migrated to the island multiple times. Although many systematic studies have investigated diverse groups of endemic species on Ulleungdo Island (Park et al., 1993; Sun and Stuessy, 1998; Weiss et al., 2002; Woo et al., 2002; Ohkawa et al., 2006; Pfosser et al., 2006; Oh et al., 2010; Shin et al., 2014), it remains unclear whether many of these species were derived from single introductions or from multiple introductions.
Chloroplast DNA is inherited by the ovulate parent in most angiosperms (Corriveau and Coleman, 1998) including Fagaceae (Dumolin et al. 1995); thus, it has been a useful tool for tracing seed dispersal (Dumolin et al., 1995; Hamilton 1999; Petit et al., 2002; Cannon and Manos, 2003; McLachlan et al., 2005). Investigation of chloroplast haplotype diversity and spatial distribution of endemic species on Ulleungdo Island would provide insight into the history of seed dispersal from mainland. One endemic species of Ulleungdo Island, Fagus multinervis Nakai, provides an excellent subject for the study of haplotype diversity. Plants of the species are an important component of the island’s forest ecosystem, as they form nearly pure stands in the deciduous forests. High genetic diversity within the population and high effective population size have been reported for the species based on allozyme data (Chung et al., 1998; Ohkawa et al., 2006).
In this study, nucleotide sequences of the psbA-trnH region of the chloroplast genome from 144 individuals of F. multinervis were determined to elucidate the diversity and spatial distribution of chloroplast haplotypes in this species and to provide insight into the origin of the species.
Materials and Methods
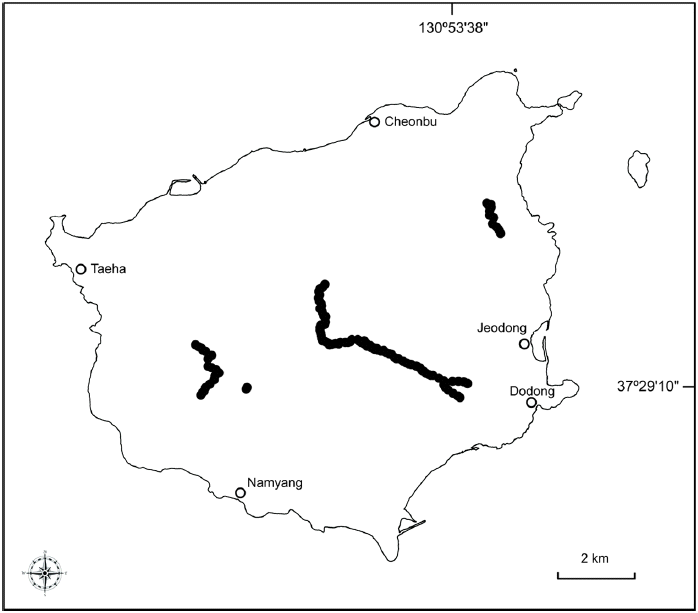
Plant materials were sampled throughout Ulleungdo Island, as plants of F. multinervis dominate the deciduous forest. Leaves were collected from 144 fully mature trees in four subpopulations (Table 1; Fig. 1). Eighty-five plants were sampled from the south-facing slope along the Seonginbong trail, and 26 plants were sampled from its north-facing slope. Nineteen samples were collected in Taeharyung and Namyang, representing the western subpopulation, and 14 in Baekwoondong, representing the eastern subpopulation. Voucher information is provided in Table 1. Species closely related to F. multinervis, namely F. crenata Blume, F. engleriana Seemen ex Diels, and F. japonica Maxim., were also included (Table 1).
The psbA-trnH region was examined because it was shown to be the most variable of the four regions of chloroplast DNA (trnK-matK, trnL-trnF, psbA-trnH, and atpB-rbcL) studied in eight Fagus species in a preliminary analysis (Oh et al., unpublished data). Total DNA was isolated from leaves, which had been dried in silica gel in the field, using the DNeasy Plant Mini Kit (Qiagen, Valencia, California, USA). The psbA-trnH region was amplified via polymerase chain reaction (PCR) using WizPure Taq DNA polymerase (Wizbio Solutions, Seongnam, Korea) in 25-μL reactions under the following conditions: initial denaturation at 95°C for 3 min, 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 7 min. The primers psbA (5GTTATGCATGAACGTAATGCTC-3) and trnH (5-CGCGCATGGTGGATTCACAATCC-3), both published by Sang et al. (1997), were used for amplification and sequencing of the psbA-trnH region. PCR products were examined in 1% agarose gels, purified, and directly sequenced in SolGent (Daejeon, Korea) using the sequencing primers. Sequences were edited in Sequencher version 5.0 (Gene Codes Corporation, Ann Arbor, MI, USA) and aligned using MUSCLE (Edgar, 2004).
Phylogenetic analysis was conducted using the maximum parsimony (MP) method. All characters were treated as unordered, and weighted equally in the MP analyses in PAUP* (Swofford, 2002). Gaps resulting from multiple alignment of indels were scored as separate characters. An exhaustive search was used to find the MP tree for the data. The bootstrap analysis (Felsenstein, 1985) with 500 pseudoreplicates was conducted with simple sequence addition and TBR branch swapping.
Results and Discussion
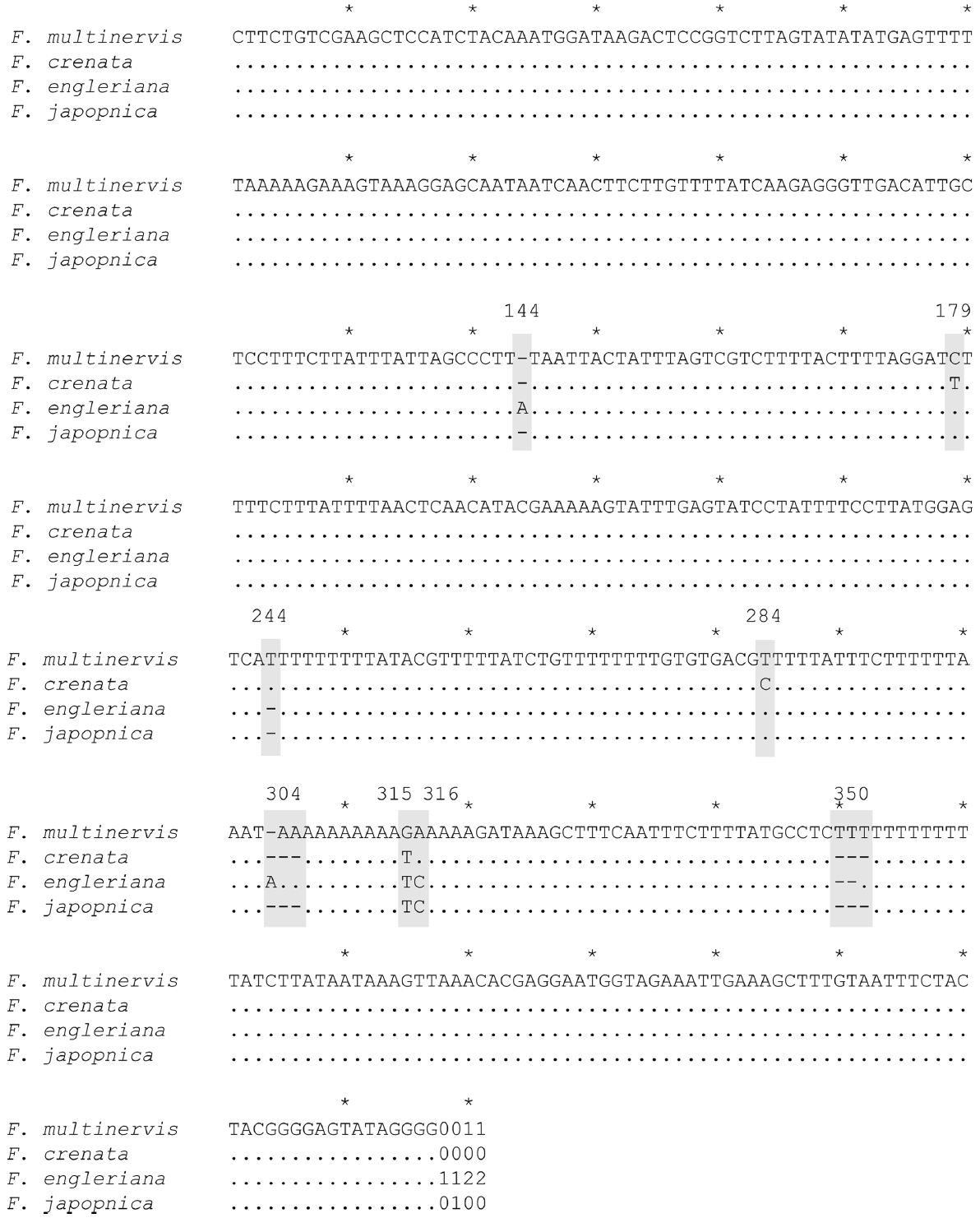
The length of the amplified psbA-trnH region ranged from 429 bp in F. japonica to 435bp in F. multinervis. The length of the region in F. crenata and F. engleriana was 430 bp and 434 bp, respectively. The nucleotide sequences were deposited in GenBank (KT382182–KT382185). The final alignment resulted in 441 sites, including four scored indel characters (Fig. 2). There were eight variable sites, including four indels. Two of the eight variable sites were parsimoniously informative, of which only one site was derived from base substitution.
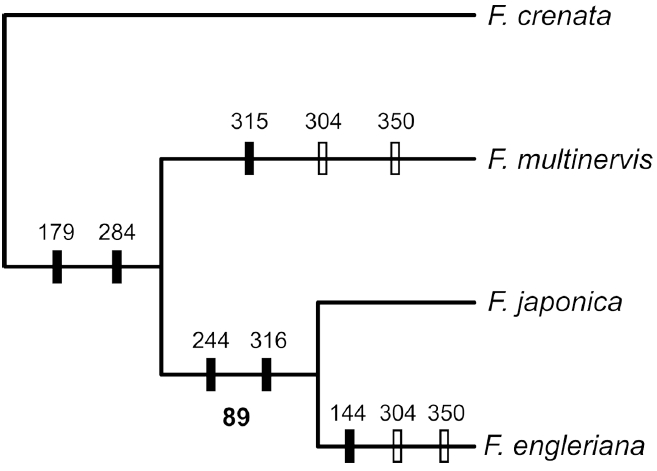
All 144 individuals of F. multinervis sampled on Ulleungdo Island had the same haplotype (Fig. 2). Fagus multinervis has a unique haplotype supported by three changes, one from a base substitution and two from indels (Figs. 2, 3). Fagus multinervis, F. japonica, and F. engleriana are members of subg. Engleriana (Shen, 1992), and previous molecular studies strongly supported the monophyly of the subgenus (Denk et al., 2005). Thus, the tree was rooted by F. crenata.
Phylogenetic analysis of the psbA-trnH data resulted in a single MP tree, with the length of 10, CI of 1.0, and RI of 1.0. The MP tree of subg. Engleriana shows that F. multinervis is sister to F. engleriana and F. japonica (Fig. 3), suggesting that the insular species should have been derived from the common ancestor that gave rise to F. multinervis and an ancestor of F. engleriana and F. japonica. Although no Fagus populations are present on the Korean Peninsula, fossil records indicated that the beech dominated temperate deciduous forests on the southeastern coast of Korea (Paik et al., 2012).
Results of this study indicate that haplotype diversity in F. multinervis is very low in comparison with other species of Fagus. In F. crenata, nine haplotypes were reported in 488 individuals by using the trnT-trnF region (Okaura and Harada, 2002). In another study that used trnL-trnF and the entire trnK intron (including matK), 13 haplotypes were found in 109 individuals of F. crenata (Fujii et al., 2002). In North American F. grandifolia, 17 haplotypes were found in 122 trees on the basis of three non-coding regions, trnL-trnF, trnK-matK, and atpB-rbcL (McLachlan et al., 2005). In a study of Greek populations of F. sylvatica, 13 haplotypes were found in 40 populations by using cpDNA microsatellite data (Hatziskakis et al., 2009). Chloroplast regions used in the previous studies, such as trnK-matK, trnL-trnF, psbA-trnH, and atpB-rbcL, were screened in a preliminary analysis for this study, and a single haplotype was found in F. multinervis. A genomic approach using a large number of samples may be necessary to completely understand the haplotype diversity of F. multinervis.
A single introduction of the seed of the progenitor of F. multinervis to the island, followed by its subsequent isolation, would have resulted in low haplotype diversity in F. multinervis. This is probably the simplest explanation for the observed results, as frequent immigration of progenitors and their establishment on the island during the early stage of evolution of F. multinervis would have led to increased haplotype diversity. In addition, a low rate of molecular evolution may be associated with the low haplotype diversity. Rates of evolution of the chloroplast genome in tree species that have long generation times, such as F. multinervis, are lower than the rates in herbaceous species that have short generation times (Smith and Donoghue, 2008).
By contrast, high genetic diversity within a population of F. multinervis was reported on the basis of allozyme analyses (Chung et al., 1998; Ohkawa et al., 2006). The mean number of alleles per locus (A) and the mean number of alleles per polymorphic locus (Ap) were as high as in other species of Fagus, such as F. sylvatica, F. grandifolia, F. japonica, and F. crenata (Ohkawa et al., 2006). The numbers of observed and expected heterozygosities were comparable to those in F. japonica and much higher than those detected in other species (Ohkawa et al., 2006). Fagus japonica is a species endemic to Japan and distributed on the Pacific side of northern Honshu, Shikoku, and Kyushu, whereas another Japanese endemic species, F. crenata, shows a wide distribution range from southern Hokkaido to Kyushu. Fagus grandifolia and F. sylvatica are distributed widely in eastern North America and throughout Europe, respectively. Ulleungdo Island is very small when compared with the large distributional range of other species. If F. multinervis was derived from a single ancestor and isolated from its progenitor, we would expect low diversity in the allozyme as well as in chloroplast data because of the founder effect. In a survey of allozyme variation in six native species on Ulleungdo Island (Chung et al., 1998), a low level of variation was found in Anemone maxima Nakai, Campanula takesimana Nakai, Tiarella polyphylla D. Don, and Thymus magnus (Nakai) Nakai, and a moderately high level of diversity was detected in F. multinervis and Pinus parviflora Siebold & Zucc.
Why does F. multinervis have a high genetic diversity in the nuclear gene pool but a very low diversity in its chloroplast haplotype? Dispersal syndromes and population structure of its progenitor on the mainland may have resulted in the pattern of diversification on Ulleungdo Island. Seeds of progenitors of native species of Ulleungdo could have been dispersed by sea current, wind, or birds from the neighboring mainland. Dispersal via a floating mass such as a broken tree trunk may have been possible, as the North Korean Cold Current and the East Korea Warm Current flow around Ulleungdo Island. However, the nut of F. multinervis is relatively large and its surfaces are smooth, and it probably would not have survived long-distance dispersal via sea currents.
The Westerlies and typhoons that blow over Ulleungdo Island may play an important role in the dispersal of propagules from the mainland to Ulleungdo Island. Beech nuts are too heavy to be transported by the Westerlies. Typhoons accompanied by strong winds, however, may have transported the nut from the mainland to Ulleungdo Island, as typhoons often move through the East Sea in August and September, the fruiting season of F. multinervis. Thus, nuts of F. multinervis may have been dispersed via typhoons. However, the probability of wind dispersal and establishment of beech nuts would be low, as it has to be assumed that a beech nut needs to be air-borne for more than several hours to reach the small island.
Dispersal of nuts via birds may also have resulted in the colonization of propagules from the mainland. It is likely that nuts of the progenitor of F. multinervis were biotically dispersed. Although bird species that disperse the nuts of F. multinervis on Ulleungdo Island are unknown, it has been widely documented that beech nuts of the North American F. grandifolia are dispersed by blue jays (Johnson and Adkisson, 1985) and those of the European F. sylvatica are carried by European jays (Perea et al., 2007). Beech forests usually form a pure stand (Hukusima et al., 2013), but chloroplast haplotype diversity within a population is generally low. Chloroplast haplotype is geographically structured in many species of Fagus and other species of Fagaceae (Whittemore and Schaal, 1991; Fujii et al., 2002; Okaura and Harada, 2002; Petit et al., 2002; Kano et al., 2004), and very few haplotypes or even a fixed haplotype is found in one population. If the nuts of the progenitor of F. multinervis were introduced frequently to Ulleungdo Island by birds that had actively harvested the nuts, a large number of seeds may have been brought to the island over time before the extinction of the progenitor on the mainland. Repeated colonization of seeds from the same progenitor population in which chloroplast DNA is geographically structured may have resulted in the diversification pattern found on Ulleungdo Island.
Alternatively, the pattern of genetic diversity of F. multinervis may have been associated with independent dispersal of seeds and pollen. In this scenario, long-distance dispersal of the seed of the progenitor of F. multinervis from the mainland via birds or typhoons to Ulleungdo Island is a rare event that resulted in the low chloroplast haplotype diversity observed in the current study. However, immigration of pollen from the mainland may have frequently occurred, which would have resulted in increasing genetic diversity in the nuclear genome. Pollination occurs in April to May, and the Westerlies may have played a role in the transport of pollen. As discussed above, fossil record indicates that beech forests were developed on the eastern coast of the Korean Peninsula until the vegetation went extinct because of volcanic activity (Paik et al., 2010; 2012). It may be possible that these populations may have been progenitors of Ulleungdo beech.
In conclusion, the patterns of genetic diversity of F. multinervis that show a relatively high allozyme diversity with a very low chloroplast haplotype diversity may be explained by frequent seed dispersal from geographically structured progenitors on the mainland or by differences in the dispersal rates of seeds and pollen from source areas. Detailed comparative phylogeographic studies of other species endemic to Ulleungdo Island and their close relatives on the neighboring mainland will be necessary for a more complete understanding of the evolution of the island’s native species.