The Korean peninsula, which is on the far east coast of Asia, has rich floristic diversity compared to its relatively small size (Park, 2005). It can be divided into three floristic regions with adjacent regions (i.e., China, Japan, and Far Eastern Russia) and possess diverse habitats due to topographic and climatic complexities (Park, 2005; Chang et al., 2011). Although large number of taxa in the Korean peninsula are shared with adjacent regions, seven genera are endemic to the Korean peninsula, including Mankyua B.-Y. Sun, M. H. Kim & C. H. Kim (Ophioglossaceae), Megaleranthis Ohwi (Ranunculaceae), Pentactina Nakai (Rosaceae), Echinosophora Nakai (Fabaceae), Abeliophyllum Nakai (Oleaceae), Hanabusaya Nakai (Campanulaceae), and Coreanomecon Nakai (Papaveraceae) (Park, 2005; Kim and Park, 2013). All these genera are monotypic, which comprises single species, except for Hanabusaya [H. asiatica (Nakai) Nakai, H. latisepala Nakai]. Recent molecular phylogenetic studies shed light on the origin of these Korean endemic genera and their status as endemic genera (e.g., Abeliophyllum, Kim et al., 2000; Echinosophora, Lee et al., 2004; Hanabusaya, Roquet et al., 2008; Mankyua, Sun et al., 2009; Megaleranthis, Kim et al., 2009; Pentactina, Lee and Hong, 2011). Echinosophora, however, is currently treated as Sophora (Lee et al., 2004). Due to continuing decline in quality and quantity of habitat, all endemic genera of Korea were classified as threatened categories of IUCN Red List Categories (Abeliophyllum, endangered [EN]; Pentactina, critically endangered [CR]; Hanabusaya asiatica, EN; Mankyua, CR; Megaleranthis, EN), except for Coreanomecon (Chang et al., 2016). Several phylogenetic and population genetic studies have been conducted for these endemics but their divergence times from their sister lineages have not been determined. This lack of estimated divergence time hinders us to fully understand the origin and evolution of endemic genera in the Korean peninsula as well as to manage and develop conservation strategy of highly threatenened and valuable floristic members in Korea.
Mankyua chejuense B.-Y. Sun, M. H. Kim & C. H. Kim (Ophioglossaceae) is the only member of the genus Mankyua and is endemic to Jejudo Island, Korea. This monotypic genus has unique combination of morphological characters which differ from other genera of Ophioglossaceae (i.e., trophophore blade, trophophore venation, sporophore, and sporangium). Since it has unique morphology and is rare and endemic to volcanic island, considerable attention has been focused on this enigmatic species. Several studies about morphology, phenology, molecular phylogenetics, conservation, and population genetics were conducted (Chung et al., 2010; Hyeon et al., 2010, 2011; Hyun et al., 2014; Kim et al., 2014; Stuessy et al., 2014). Although previous phylogenetic studies have examined the relationships within Ophioglossaceae (Sun et al., 2009; Shinohara et al., 2013), phylogenetic position of Mankyua is still controversial.
Jejudo island is of volcanic origin and 90km south off the coast of the Korean peninsula (Woo et al., 2013). While the family Ophioglossaceae is early diverged basal lineage of monilophytes group, Jejudo Island is very young with the age of approximately 2 million years old (Woo et al., 2013). Since Jejudo Island has recurrently connected to adjacent continents during the glacial cycles in the Quaternary Period, the flora of Jejudo Island has been affected by the various floristic elements from the Korean peninsula, China, and Japan, where habitats are diverse. Despite continuous interests on this highly enigmatic old lineage of Ophioglossaceae, there has been no attempt to estimate the divergence time of Mankyua, specifically for its split from the closest lineage and the crown age. Therefore, in the present study, we explored the phylogenetic position of Mankyua within Ophioglossaceae and estimated the divergence time of M. chejuense.
Materials and Methods
DNA sequence and phylogenetic analyses
All DNA sequences were retrieved from GenBank (Appendix 1). The rbcL and matK sequences were aligned using CLUSTAL W (Larkin et al., 2007), implemented in Geneious version 7.1.7 (Kearse et al., 2012). The alignment was further examined and slightly edited manually as necessary. For the combined cpDNA datasets, we selected six taxa of two genera, Huperzia and Isoetes, as outgroups (Pryer et al., 2004). A total of 42 accessions for 37 taxa were used for phylogenetic analysis. The combined data set was analyzed by maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI) method. The MP phylogenetic analyses was performed using PAUP* version 4.0b10 (Swofford, 2002). Characters were treated as unordered and all character transformations were weighted equally. The MP analyses were conducted using heuristic search options with simple stepwise addition of taxa, tree-bisection-reconnection branch swapping, and saving multiple trees. Bootstrap values (Felsenstein, 1985) were calculated from 1,000 replicate analyses using the same heuristic search options as above. Gaps were treated as missing data. For the ML and BI analysis, the best-fit model was selected based on the Akaike information criterion implemented in the program jModelTest version 2.1.6 (Darriba et al., 2012). For both ML and BI analysis, GTR + I + G model was selected. ML analyses were conducted using RAxML 8.0.26 (Stamatakis, 2014) implemented in raxmlGUI version 1.3.1 (Silvestro and Michalak, 2012). ML trees were calculated using 1,000 rapid bootstrap inferences. BI analysis was performed with 5,000,000 generations initiated with a random starting tree, sampling every 500 generations and allowing the program to estimate the likelihood parameters required. We discarded 25% of the samples as burn-in.
Divergence time estimation
Divergence times of the family Ophioglossaceae and major lineages within the family were estimated by BI approach using the program BEAST v.2.3.1 (Bouckaert et al., 2014). Huperzia and Isoetes were excluded from the analysis to reduce the computational burden. As earlier study of molecular dating of ferns (monilophytes (Pryer, 2014)), we used four fossil calibration points to estimate the divergence time of Ophioglossaceae. We incorporated four fossil constraints from a reassessment of the fern fossil record and the root of the resulting tree was used as a calibration point based on the concurrent appearance of fossils belonging to each of these lineages in the Middle Devonian. We employed a relaxed molecular clock model (Drummond et al., 2006) relying on uncorrelated rates drawn from a log-normal distribution, and a Yule tree prior for speciation. Two independent Markov chain Monte Carlo runs were performed with 5,000,000 generations each, with every 500 generation sampled. A burnin of 10% per run was discarded after assessing convergence with Tracer version 1.4.1. To obtain an estimate of the phylogenetic tree with mean divergence time and 95% highest posterior density (HPD) intervals, the program TreeAnnotator v.1.7.5 was used as it summarizes the post burn-in trees and their parameters.
Results
Phylogenetic position of Mankyua
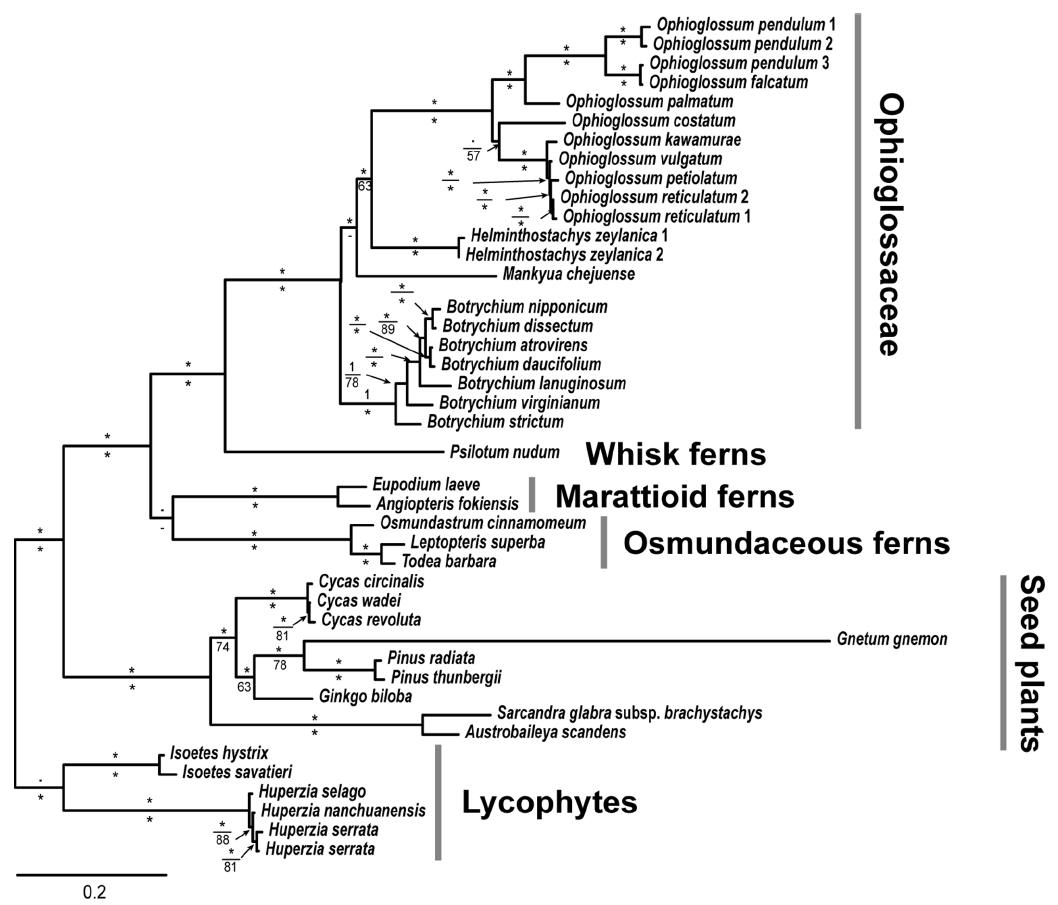
The combined cpDNA data set included a total of 42 accessions. A total 2,032 aligned sites for concatenated cpDNA characters were used for MP, ML, and BI analyses. Of the 2,032 characters, 836 were constant, 123 were variable but parsimony-uninformative, and 1,073 were parsimony-informative characters. The MP analysis found one most parsimonious tree (Fig. 1) with a tree length of 4,350, a consistency index of 0.4570 (0.4388 excluding uninformative characters), and a retention index of 0.7862.
Since some topological incongruences were found among ML/BI and MP trees, our results and discussion are based on ML (Fig. 2) and MP (Fig. 1) trees. As shown in both MP and ML trees, the family Ophioglossaceae is monophyletic with strong support value (posterior probability [PP] 1, ML bootstrap support [BS] 100%, MP BS 100%). In addition, the monophyly of three genera (Botrychium, Ophioglossum, and Helminthostachys) is strongly supported (PP 1, ML BS 100%, MP BS 100%). In ML and BI trees, Botrychium was resolved as the earliest diverged lineage of Ophioglossaceae clade (PP 1, ML BS 100%) and showed sister relationship with the clade containing remaining three other genera in Ophioglossaceae (Fig. 2). Mankyua shared its most recent common ancestor (MRCA) with Helminthostachys and Ophioglossum (PP 0.92, ML 46%). However, MP tree demonstrated that Mankyua was resolved as the earliest diverged lineage in Ophioglossaceae (MP BS 69%) (Fig. 1).
Divergence time estimation
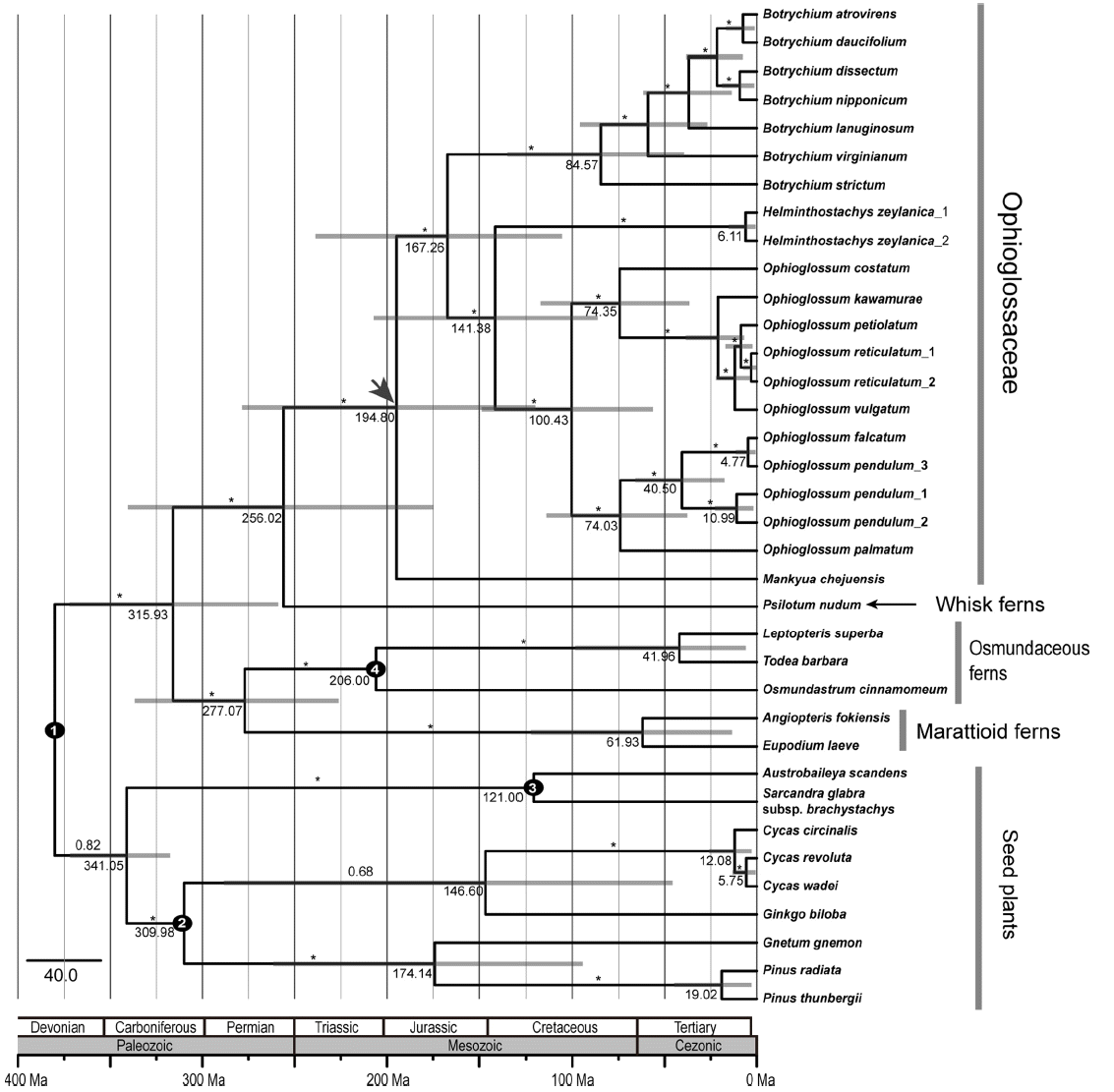
To estimate the ages for major clades within Ophioglossaceae, we conducted Bayesian analyses using BEAST to obtain 95% HPDs for the nodes of interest. The maximum-credibility tree based on the relaxed molecular clock analysis of Ophioglossaceae using four fossil calibration points is shown in Fig 3. The stem and crown age of Ophioglossaceae was estimated to be 256 Ma and 195 Ma, respectively (during the late Permian and Triassic) (Fig. 3). Mankyua was the earliest lineage diverged during the early Jurassic (195 Ma), with the MRCA of three genera Botrychium, Helminthostachys, and Ophioglossum. Botrychium diverged from the remaining Helminthostachys + Ophioglossum lineages approximately 167 Ma, with Helminthostachys and Ophioglossum splitting from one another during the early Cretaceous (141 Ma) (Fig. 3). Ophioglossum subsequently diverged into two lineages in the Mid Cretaceous (100 Ma) (Fig. 3). The MRCA of Botrychium and Ophioglossum was estimated to be 85 Ma and 100 Ma, respectively.
Discussion
The phylogenetic position of Mankyua within Ophioglossaceae clade in MP and ML/BI analyses differs significantly. The ML and Bayesian trees (Fig. 2) indicate that Botrychium was resolved as the earliest-diverging lineage of Ophioglossaceae, and that Mankyua is the sister to the clade containing Ophioglossum and Helminthostachys with low ML bootstrap support. In the MP tree (Fig. 1), however, Mankyua was diverged first in Ophioglossaceae clade and Botrychium and Helminthostachys + Ophioglossum showed sister relationship with moderate bootstrap support (MP BS, 69%). In the previous studies, Sun et al. (2009) presented MP and neighbor joining (NJ) trees of Ophioglossaceae using rbcL data and showed that Ophioglossum diverged first and sister to the clade containing Mankyua, Botrychium and Helminthostachys. Mankyua showed sister relationship with the clade containing Botrychium and Helminthostachys with low bootstrap support. Shinohara et al. (2013) pointed out that MP and NJ approaches are sensitive to long-branch attraction (Sanderson et al., 2000; Anderson and Swofford, 2004), supporting the ML tree topology based on the rbcL and matK data set. According to the result of Shinohara et al. (2013), Mankyua is the earliest-diverging lineage in Ophioglossaceae clade and Botrychium was sister to the clade containg Helminthostachys and Ophioglossum with moderate support value. Although the same data sets of Shinohara et al. (2013) and more outgroup taxa were analyzed in our study, phylogenetic position of Mankyua was different from that of Shinohara et al. (2013), providing no concrete position of Mankyua.
In terms of morphological characteristics, Mankyua and Helminthostachys shares major characteristics associated with vegetative organ (i.e., creeping rhizomes, alternate leaf arrangement, and ternately compound trophophore blade) and reproductive organ morphological characteristics of Mankyua are shared with Ophioglossum (i.e., linear and fleshy sporophore, sunken and horizontally dehiscent sporangium), suggesting Mankyua is much resemble with Helminthostachys and Ophioglossum rather than Botrychium (Sun et al., 2001). Shinohara et al. (2013) reported the chromosome number of Mankyua (x = 130), the highest base chromosome number in Ophioglossaceae, and compared with other genera in Ophioglossaceae. While outgroups (Angiopteris and Psilotum) have low base chromosome numbers of x = 40 and 52, respectively, the most genera of Ophioglossaceae have high base chromosome numbers (Ophioglossum, x = 120; Helminthostachys, x = 94) but Botrychium (x = 44, 45, and 46) (Shinohara et al., 2013). Thus, they suggested that the initial chromosome number of this family was seemed low and increased through polyploidization events, which are common in ferns and lycophytes. Despite several previous studies, we still have room to carry out more detailed morphological, cytological and molecular phylogenetic studies to determine precise phylogenetic position of Mankyua and to resolve phylogenetic relationships among major lineages within Ophioglossaceae.
For the first time, we estimated divergence time of Korean endemic and monotypic genus Mankyua chejuense. Molecular dating tree showed no major topological incongruence with the ML tree except Mankyua chejuense and Ginkgo biloba (Figs. 2, 3). The family Ophioglossaceae diverged during the Permian (256 Ma), the late Paleozoic, with a crown age of 195 Ma, the late Jurassic in Mesozoic (Fig. 3). In the previous study, Pryer et al. (2004) estimated the divergence time and origin and diversification of major fern clades including Ophioglossaceae. According to the results of their study, the origin of Ophioglossaceae was estimated to be 306 Ma, the late Carboniferous, in the Paleozoic, with a crown age of 161 Ma in the Jurassic, the mid Mesozoic era. Since we have incorporated more extensive taxon sampling of Ophioglossaceae, we suggest that our current results may reflect more accurate divergence time of Ophioglossaceae.
Our molecular dating tree (Fig. 3) indicated that Mankyua shared the MRCA with Botrychium, Ophioglossum, and Helminthostachys in approximately 195 Ma, the early Jurassic period in the Mesozoic era. Mankyua chejuense is the only surviving member of genus Mankyua, basal lineage of monilophytes. This ancient relic species, M. chejuense, is distributed in highly restricted area (5.7 km × 4 km), northeastern part of young Jejudo Island, dominated by Ulmus, Cudrania, Camellia, Ligustrum, and Rosa species with small population sizes (Kim, 2004; Chung et al., 2010; Hyeon et al., 2010, 2011). Previous population genetic study of Mankyua with allozyme markers (Chung et al., 2010) revealed extremely lower levels of genetic diversity (HES = 0.007, HEP = 0.005) than other Korean endemic genera (Abeliophyllum distichum, HES = 0.143, HEP = 0.110, Chung, 1999; HES = 0.178, HEP = 0.13, Kang et al., 2000; Hanabusaya asiatica, HES = 0.217, Chung et al., 2001; Megaleranthis saniculifolia, HES = 0.088, Chang et al., 2005; HES = 0.151, HEP = 0.083, h = 0.135 [intersimple sequence repeat, ISSR], Jeong et al., 2010). In addition, Chung et al. (2010) found that high degree of popualtion differentiation in Mankyua chejuense and suggested that founder effect and intragametophytic self-fertilization contributed to such pattern of genetic diversity and differentiation. Very surprisingly, we found similar genetic patterns in another monotypic genus, Lactoris (Lactoridaceae), which is also considered the remnant of an old angiosperm lineage and endemic to the Juan Fernández Archipelago (ca. 4 million years old). Comprehensive population genetic studies are conducted with various molecular markers (i.e., ribosomal DNA length, randomly amplified polymorphic DNA, allozyme, and ISSR), but lack of or low genetic diversity was found within populations (Brauner et al., 1992; Crawford et al., 1994, 2001). Furthermore, Crawford et al. (2001) found over 73% of the ISSR diversity occurred across populations, suggesting very high degree of population differentiation. For the major factors responsible for this genetic patterns, founder effect, genetic drift associated with small population sizes, and a selfing breeding system have been suggested. To better understand for Mankyua chejuense, comprehensive studies of morphology, ecology, breeding system, and population genetics with other highly variable and informative molecular markers would be rewarding. In addition, it would be of great value to estimate the origin and divergence time of other Korean endemic genera to have better picture on these valuable floristic elements in the Korean peninsula.