A fern genus Hypodematium Kunze is characterized by short rhizomes and swollen bases of stipes densely covered with scales; stipes and lamina usually with acicular long hairs, mixed glandular hairs or glabrous; largest basal pinna; and chromosome number x = 40 and 41, occurring limitedly only on limestone rocks (Shing et al., 1999; Zhang and Iwatsuki, 2013; Wang et al., 2014). Firstly, Iwatsuki (1964) recognized the genus with four species and one subspecies. It currently consists of 18 species distributed in rock crevices of limestone areas mostly in subtropical and warm-temperate regions of Asia and Africa, with high species diversity in China (12–14 species) (Shing et al., 1999; Zhang and Iwatsuki, 2013; Wang et al., 2014). Hypodematiaceae firstly suggested by Ching (1975) including the genus Hypodematium and two other genera, Didymochaena and Leucosegia. It has been suggested that Hypodematiaceae is a monophyletic family only with Hypodematium based on molecular study (Tsutsumi and Kato, 2006; Liu et al., 2007; Schuettpelz and Pryer, 2007; Christenhusz et al., 2011).
Three Hypodematium species are native to Korea. Formerly, only one Hypodematium species, H. glanduloso-pilosum (Tagawa) Ohwi, has been recognized for a long time (Park, 1975; T. B. Lee, 1980; Y. N. Lee, 2006; Korea National Arboretum, 2008; Lee and Lee, 2015; Kim and Sun, 2015), and occurs widely in limestone or rock crevices throughout the country except Jejudo Is. in Korea. However, we have recently reported one unrecorded taxon, H. squamuloso-pilosum Ching, found on rocks at a limestone mountain in Yeongwol, Gangwon-do, and one new taxon, H. angustifolium, observed in Okcheon, Chungcheongnam-do (Lee et al., 2017; Lee and Lee, 2018). Traditionally, these taxa of Hypodematium can be distinguished based on vegetative characters such as pinnatifid lamina, glandular hairs on indusia, and narrower or linear lanceolate scales on stipe bases.
Among the three Korean Hypodematium, H. glanduloso-pilosum is distributed in the central and southern area of China, South Korea, Japan and Thailand (Iwatsuki, 1995; Zhang and Iwatsuki, 2013). The species has broad variation in morphological characters such as leaf segregation and distribution of glandular hairs and scales, position of sporangia and indusia. H. squamuloso-pilosum occurs in central and southern area of China and restricted northern area of South Korea (Iwatsuki, 1995; Zhang and Iwatsuki, 2013; Lee et al., 2017). In addition, H. angustifolium exhibits a restricted distribution as an endemic taxon to Korea, which is recently described as a new species with characteristics of linear lanceolate scales with shortly or lengthily serrate margins on rhizomes and base of stipes, thinner and longer lamina, almost glabrous stipes, oblong triangular basal pinna, and almost glabrous indusia (Lee and Lee, 2018).
Chromosome numbers of several species has been reported with aneuploidy and polyploidy in the genus, but the two basic chromosome numbers of Hypodematium are known. H. glanduloso-pilosum and H. squamuloso-pilosum are n = 41 (x = 41, plesiomorphic), and H. fordii is n = 40 (x = 40) considered as apomorphic character state. H. crenatum is a tetraploid with chromosome number of n = 82 (Wang et al., 2014). Genome size correlates with chromosome number across all ferns despite some substantial variation in both traits (Clark et al., 2015).
Although the genus is small with about 18 species, diverse character variation has been observed in morphology, geography, and cytology. Broad ranges of morphological and geographical characters make hard to determine species delimitations and taxonomic treatments. The monophyly of the genus and phylogenetic relationships among taxa within the genus have not been clarified using molecular data.
Documentation of intraspecific variations in cpDNA has become increasingly common (Wagner et al., 1987; Soltis et al., 1991, 1992; Terry et al., 2000). Consideration of data from chloroplast DNA markers are important because any significant discordance of relationships between data sets may serve to identify past hybridization (Doyle, 1992; Rieseberg and Brunsfeld, 1992). In cases where a species is polymorphic cpDNA is diagnostic for a second species, interspecific hybridization is a plausible explanation. Concordance in the geographic distribution and relationships suggested by independently evolving characters (e.g., cpDNA) provides additional supports (Terry et al., 2000).
In the present study, we attempted to investigate the following questions: (1) clarify the monophyly of the genus Hypodematium and each species, especially Korean taxa, H. glanduloso-pilosum, H. angustifolium and H. squamuloso-pilosum (2) identify major lineages within the genus, (3) clarify morphological variation range of Korean taxa, and (4) test traditional classification in a phylogenetic framework. This is a first attempt to evaluate the phylogenetic relationships among Hypodematium taxa. Morphological and geographical character variation in the genus Hypodematium is discussed within a molecular phylogenetic framework.
Materials and Methods
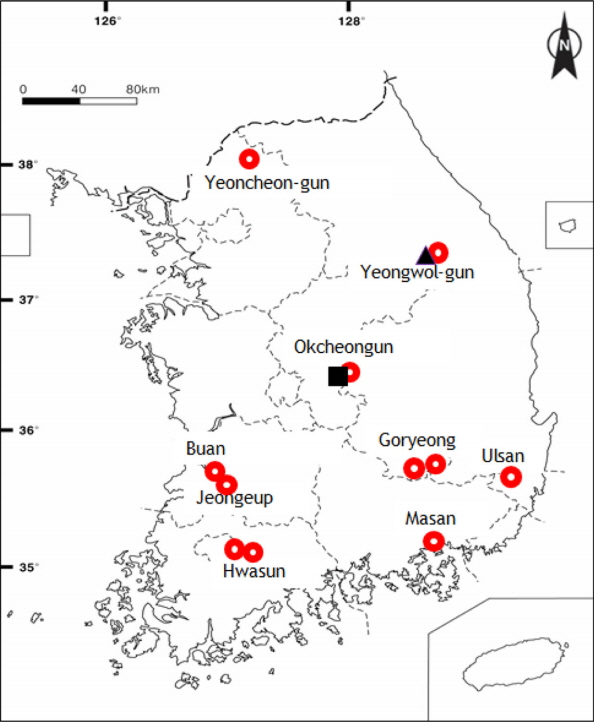
The sources of plant materials for morphology and DNA analysis and GenBank accession numbers are listed in Appendix 1. All voucher specimens were deposited at Ewha Womans University Herbarium (EWH). To evaluate H. glanduloso-pilosum and related taxa of Hypodematium, we collected from 9 populations (Yeongwol-gun, Gangwon-do; Yeoncheon-gun, Gyeonggi-do; Okcheon-gun, Chungcheongbuk-do; Buan, Jeongeup, Jeonlabuk-do; Hwasun, Jeollanam-do; Goryeong, Gyeongsangbuk-do; Ulsan, Masan, Gyeongsangnam-do) of Korea to maximize their geographic variation, habitat differences, and morphological diversity (Fig. 1). We also sampled three individuals of H. squamuloso-pilosum and six of H. angustifolium from Korea as the presumably closely related out-group taxa (Appendix 1). Fourteen morphological characters were observed and measured from mature individuals, including H. glanduloso-pilosum and related taxa referred by Iwatsuki (1992), Wang et al. (2010), and Zhang and Iwatsuki (2013). In comparing the molecular analysis, we got from DNA bank nine accessions containing one of H. glanduloso-pilosum and one of H. fordii from Japan in Hypodematium, and included one of Didymochlaena truncatula (Sw.) J. Sm. and Dryopteris maximowiczii (Baker) Kuntz. for outgroup. For most populations, one to six individuals were examined in cpDNA and morphological analyses. A total of 28 accessions from eight taxa of Hypodematium, including two accessions from the two outgroup taxa were used for cpDNA, rbcL analysis. In addition, 20 accessions of Hypodematium, including the two outgroups, were studied for psbA-trnH. The rbcL sequences for 19 accessions and psbA-trnH sequence data for 15 accessions were newly analyzed (Appendix 1), and outgroup and several ingroup data (17 accessions) were obtained from GenBank. In the combined analyses of two cpDNA, the compression per each population region of each taxon were applied.
Total genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA) following the manufacturer’s instructions. Polymerase chain reaction (PCR) amplification, purifications, and DNA sequencing strategies followed White et al. (1990), according to the PCR conditions described by Lee and Downie (2006) and Lee et al. (2006).
Primers used for amplification and sequencing were rbcL-TKT-F1 and rbcL-TKT-R3N-2 for the rbcL gene (Ebihara et al., 2005), and psbA and trnHR for psbA-trnH intergenic spacer (Sang et al., 1997). Amplification was using a PTC-100 thermal cycler (MJ Research, Watertown, MA, USA). Automated sequencing analysis was performed using a sequencer (Base station, MJ Research). Chloroplast DNA regions were each aligned with gap adjustments, using the Clustal X program, followed by manual adjustment (Thompson et al., 1997). Each of and combined matrices of the two cpDNA regions were analyzed using Maximum Parsimony (MP) by PAUP*(Swofford, 2002).
Results and Discussion
Morphological analysis
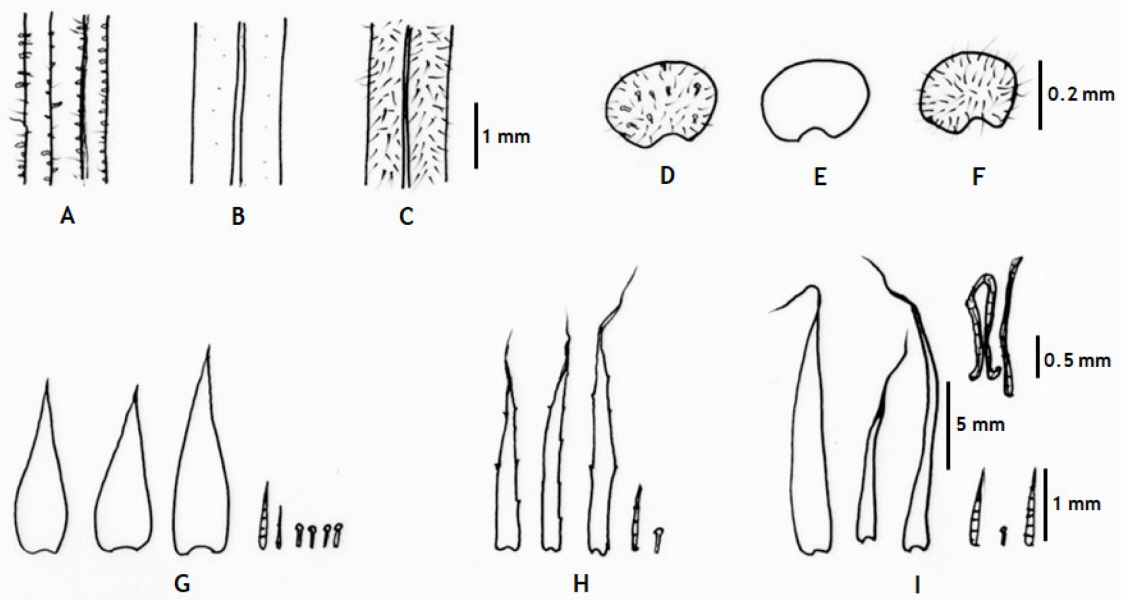
Hypodematium glanduloso-pilosum, H. angustifolium, and H. squamuloso-pilosum show variation in pinnules shapes, scale and hair shapes and presence on rhizomes and stipe bases; and hair types and distributions on stipes and indusia (Figs. 2, 3), and their major morphological characters and geographic distribution is summarized in Table 1.
H. glanduloso-pilosum was observed widely from nine populations of Korea (Appendix 1). This taxon is distributed throughout South Korea excluding Jeju-do Isl. (Fig. 1), and southern area of China, Japan, and Thailand. Its morphological variations include the broad ranges in the morphology of stipe lengths (4–27 cm), indusium positions from the middle of veinlet to near costules of pinnules, and the distribution of hairs from densely covered with white, short hairs and golden, rod-shaped glandular hairs, and segmentations of pinnule margins (Fig. 2). However, H. glanduloso-pilosum is clearly distinguished from H. angustifolium and H. squamuloso-pilosum by the characters of much wider scales (oblong lanceolate or lanceolate), dense white short hairs and golden rod-shaped glandular hairs in stipes, lamina and indusia, and broadly ovate-oblong shaped basal pinna (Fig. 3, Table 1).
The newly reported species in Korea, H. angustifolium C. S. Lee & K. Lee (Lee and Lee, 2018), is distributed in Okcheon, Chungcheongnam-do, and has distinct characters of slightly serrate scale margins, glabrous stipes; narrower lamina, pinna, and pinnules compared to the other two Hypodematium species of Korea (H. glanduloso-pilosum and H. squamuloso-pilosum). Morphologically, the new species resembles a Chinese endemic species, H. sinense, sharing short, creeping rhizomes, almost glabrous stipes, almost glabrous adaxial and abaxial surfaces of lamina, and much dense glandular hairs along rachis, costae, and veins (Zhang and Iwatsuki, 2013; Lee and Lee, 2018), but narrower lanceolate, sparsely short hairs in lamina, brown or light brown scales on rhizomes and stipe bases, short hairs, gradually over long narrow apex, narrower pinna, pinnules, and segments are only observed in H. angustifolium not in H. sinense. Moreover, the species forms a clade with H. fordii (Japan and China) and Korean taxa (Figs. 4, 5). H. angustifolium has stipe mostly almost glabrous only with a few scales with slightly or lengthily serrate margins at the base of stipes, sparsely short hairs and densely golden rod-shaped glandular hairs in lamina and lanceolate or linear lanceolate pinnules. Therefore, morphological data support H. angustifolium as an independent species and a Korean endemic.
Hypodematium squamuloso-pilosum has been considered as a Chinese endemic species until some native populations of the species were found in Korea recently (Lee et al., 2017). The species is morphologically similar to H. hirsutum in the shapes of lamina (ovate oblong) and basal pinnule (oblong to ovate oblong), presence and distribution of long hairs and scales in stipes and lamina. The finding of native populations in Korea expands distribution range of the species, and merit further biogeographic study of the species to understand speciation of the taxon.
Molecular analysis
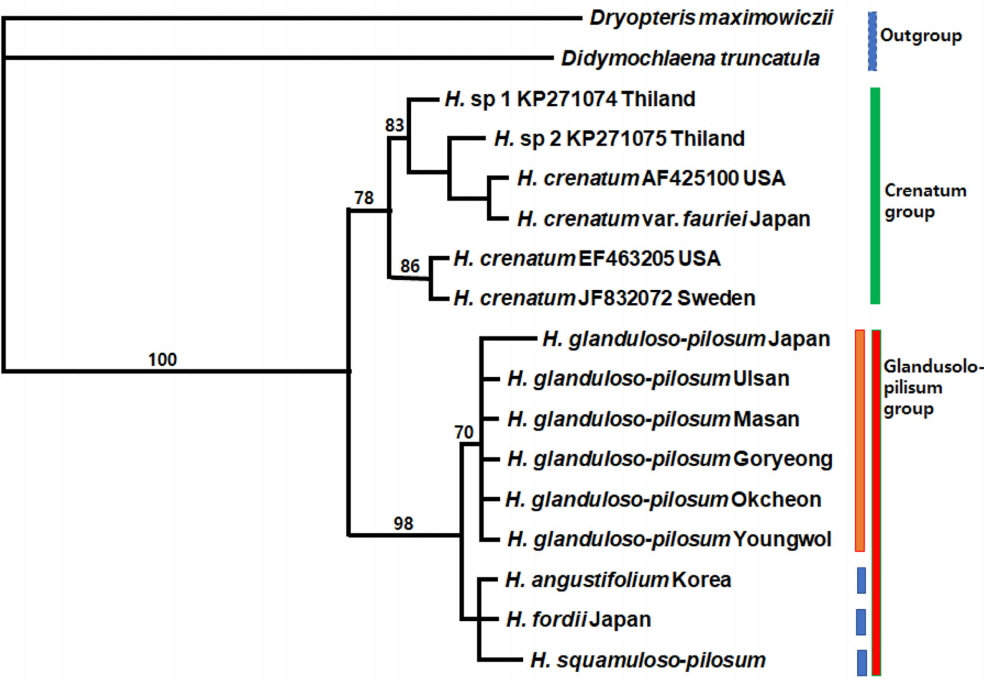
All trees result in a strongly-supported monophyletic Hypodematium clade comprising two major lineages by 100% bootstrap value (Figs. 4, 5). Of these chloroplast genes (combined 1,385 bp), the aligned length of rbcL region is 938 bp, and the aligned length of psbA-trnH IGS region is 447 bp. Of the 938 aligned positions of rbcL region, 842 sites (89.7%) are identical, 55 sites (5.9%) are parsimony uninformative, and 41 sites (4.4%) are phylogenetically informative. MP analysis of the entire rbcL sequences finds equally most parsimonious trees with a tree length (TL) of 103, a consistency index (CI) of 0.9417 (0.8723) excluding uninformative characters), and a retention index (RI) of 0.9310. One of 103 equally most parsimonious trees is shown in Fig. 4; overall, branches are moderately supported with relatively short branch lengths. This rbcL region analysis also indicates that the three species in Korea are in the same clade with Japanese H. fordii from the other species. In the rbcL region examined, 11 accessions of 6 populations of H. glanduloso-pilosum with H. angustifolium and H. fordii have no autapomorphies. Otherwise, 3 accessions of H. squamuloso-pilosum form a monophyletic group by one autapomorphy in the sequences within the glandulo-pilosum clade.
A total of 1,385 aligned sites from the combined cpDNA data set (rbcL and psbA-trnH) were used for MP analyses. Of the 1,385 characters, 1,225 are constant, 106 are variable but parsimony-uninformative, and 54 are parsimony informative characters. The MP analysis found one most parsimonious tree (Fig. 5) with a TL of 169, a CI of 0.9645 (0.9000, excluding uninformative characters), and a RI of 0.9368.
Topology of combined cpDNA tree is similar to rbcL gene tree. In the combined cpDNA data analysis, 6 populations of H. glanduloso-pilum form the monophyletic group, especially 5 populations collected from Korea. H. angustifolium, H. fordii, and H. squamuloso-pilosum are monophyletic within a Glanduloso-pilosum clade. The H. glanduloso-pilum clade is grouped with H. angustifolium, H. fordii, and H. squamuloso-pilosum, forming the Glanduloso-pilum clade with a strong support value (98% bootstrap value) and 5 substitution events.
The two clades formed in the both separate and combined cpDNA analysis are also supported by morphological characters. The Glanduloso-pilosum group (H. glanduloso-pilum + H. angustifolium + H. fordii + H. squamuloso-pilosum) has glandular or multicellular hairs, and the presence of only long hairs (without glandular hairs) is synapomorphic for the Crenatum group (H. crenatum + H. crenatum var. fauriei).
Maximum intraspecific sequence divergence values for H. glanduloso-pilosum in Korea (5 accessions) and Japan (1 accession) are 0.2%, but them for 5 populations of H. glanduloso-pilosum in Korea are all same as 0%. Maximum interspecific sequence divergence values between H. glanduloso-pilosum and H. fordii, H. angustifolium and H. squamuloso-pilosum are very low as 0.073–0.292, 0.292, and 0.147–0.367, respectively. From these results, we might hypothesize that H. squamuloso-pilosum, H. angustifolium, and H. fordii are varieties of H. glanduloso-pilosum. The Korean endemic taxon H. angustifolium, recently described as a new species (Lee and Lee, 2018), is almost homologous to H. fordii based on cpDNA data. Further study on Chinese and Japanese populations of some taxa of Hypodematium is needed to confirm it.
Maximum intraspecific sequence divergence values for H. crenatum Maximum (3 accessions) obtained from DNA bank in Thailand, USA, Sweden, and Japan are various ranging 0–2.046%. However, interspecific sequence divergence values for H. crenatum, H. sp 1, H. sp 2, H. crenatum var. fauriei nested in Crenatum clade are 0.74–1.971. Therefore, we need to reconsider their taxonomic delimitations.
Preliminary analyses of flow cytometry showed that H. angustifolium has the same level of ploidy with H. glanduloso-pilosum and H. squamulroso-pilosum (pers. observ.). H. glanduloso-pilosum and H. squamulroso-pilosum are known as diploids (Wang et al., 2014), and none of individuals with high morphological variation exhibit variation in a ploidy level. The analyses suggest that the Korean endemic taxon, H. angustifolium, is not hybrid origins.
Although the morphological characters such as hair or scale types, glandular or multicellular hairs or scales provide critical information for species delimitations, the individuals with broad variation in the morphological characters of H. glanduloso-pilosum are not supported by molecular data (rbcL and psbA-trnH, flow cytometry). Moreover, our analyses propose that H. squamuloso-pilosum (China and Korea), H. angustifolium (Korea), and H. fordii (China and Japan) were belong to the same Glanduloso-pilosum clade with H. glanduloso-pilosum (China, Korea, and Japan).
In conclusion, morphological characters distinguish each species in the genus although some character variations are high within some species, but low levels of molecular differentiation among taxa suggest reconsideration of species status. To clarify taxonomic status of each species and phylogenetic relationships among taxa, further study with broader taxon and character samplings are preferred.