/home/virtual/kjpt/journal//../xmls/kjpt-52-3-156.xml
PARK and XI: The complete chloroplast genome of Campsis grandiflora (Bignoniaceae)
Abstract
Campsis grandiflora (Thunb.) K. Schum is an ornamental species with various useful biological effects. The chloroplast genome of C. grandiflora isolated in Korea is 154,293 bp long (GC ratio: 38.1%) and has four subregions: 84,121 bp of large single-copy (36.2%) and 18,521 bp of small single-copy (30.0%) regions are separated by 24,332 bp of inverted repeat (42.9%) regions including 132 genes (87 protein-coding genes, eight rRNAs, and 37 tRNAs). One single-nucleotide polymorphism and five insertion and deletion (INDEL) regions (40-bp in total) were identified, indicating a low level of intraspecific variation in the chloroplast genome. All five INDEL regions were linked to the repetitive sequences. Seventy-two normal simple sequence repeats (SSRs) and 47 extended SSRs were identified to develop molecular markers. The phylogenetic trees of 29 representative Bignoniaceae chloroplast genomes indicate that the tribe-level phylogenic relationship is congruent with the findings of previous studies.
Keywords: Campsis grandiflora, ornamental plant, chloroplast genome, simple sequence repeats, intraspecific variations, Bignoniaceae
INTRODUCTION
Genus Campsis Lour. consists of only two species, Campsis grandiflora (Thunb.) distributed in East Asia and Campsis radicans (L.) Bureau found in North America ( Wen and Jansen, 1995). Because of its disjunct distribution, this genus was considered as a material to understand their evolutionary history, resulting that both species was estimated to be diversified at 24.4 million years ago ( Wen and Jansen, 1995). Campsis grandiflora has been utilized as an ornamental species because of their trumpet shape flowers ( Jia et al., 2012). Moreover, C. grandiflora was known to have various biological effects ( Yu et al., 2015; Oku et al., 2019), such as anti-oxidative and anti-inflammatory ( Cui et al., 2006), and useful phytocompounds ( Jin et al., 2005; Kim et al., 2007; Han et al., 2012) including triterpenoids ( Kim et al., 2005). To understand intraspecific variations of C. grandiflora chloroplast genome together with the previously published chloroplast genome isolated in China ( Chen et al., 2022), we completed the chloroplast genome of C. grandiflora isolated in Korea.
MATERIALS AND METHODS
Plant material
The sample was collected on Gangseo postal office, Seoul, Korea (37.565175N, 126.840624E). A specimen was deposited at the InfoBoss Cyber Herbarium (IN) under the voucher number, IB-01065. No permission is required for the collection.
DNA extraction and chloroplast genome determination
Total DNA was extracted from the fresh leaves using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The sequencing library was constructed using an Illumina TruSeq Nano DNA Library Preparation Kit (Illumina, San Diego, CA, USA) following the manufacturer’s recommendations with approximately 350-bp DNA fragments. 4.15-Gbp raw sequences were obtained using NovaSeq6000 at Macrogen Inc., Korea, and were filtered by Trimmomatic v0.33 ( Bolger et al., 2014). The chloroplast genome was de novo assembled with Velvet v1.2.10 ( Zerbino and Birney, 2008), and gaps were closed using GapCloser v1.12 ( Zhao et al., 2011). The genome sequence was confirmed by aligning all raw reads against the assembled genome using BWA v0.7.17 and SAMtools v1.9 (Li et al., 2009; Li, 2013). All processes were conducted in the Genome Information System ( http://geis.infoboss.co.kr) utilized in previous studies ( Kim et al., 2019e, 2020; Park et al., 2019c; Park and Xi, 2021). Geneious Prime v2020.2.4 (Biomatters Ltd., Auckland, New Zealand) was used for annotation based on the Tecomaria capensis chloroplast genome (GenBank accession number: NC_037462) ( Fonseca and Lohmann, 2018). A circular map of C. grandiflora chloroplast genome was drawn using OGDRAW v1.31 ( Greiner et al., 2019). Large single-copy (LSC), small single-copy (SSC), and inverted repeat (IR) regions were determined by bl2seq ( Tatusova and Madden, 1999).
Identification of intraspecific variations
Single nucleotide polymorphisms (SNPs) and insertions and deletions (INDELs) were identified from the pair-wise sequence alignment of the two C. grandiflora chloroplast genomes conducted by MAFFT 7.450 ( Katoh and Standley, 2013) with ‘Find variations/SNPs’ implemented in Geneious Prime 2020.2.4 (Biomatters Ltd.), which has been used in the previous studies investigating intraspecific variations on organelle genomes ( Kim et al., 2021a; Oh et al., 2021; Suh et al., 2021). INDEL region was defined as the continuous INDELs.
Identification and comparative analysis of simple sequence repeats
Simple sequence repeats (SSRs) were identified on the chloroplast genome sequence using the pipeline of the SSR database (SSRDB; http://ssrdb.infoboss.co.kr/) which has been utilized in several organelle genomic studies ( Lee et al., 2020; Choi et al., 2021; Park et al., 2021d, 2022). The SSR is conventionally recognized as the nucleotide array composed of repeats with one or up to six base pair units. For example, monoSSR refers an array of nucleotide repeats containing a particular base and hexaSSR an array of nucleotide repeats containing six base pair unit. The overall length of SSR is mostly over 10 base pairs. In this study, we tried to classify SSR with more criteria which has been applied in previous analyses ( Gandhi et al., 2010; Chen et al., 2015; Cheng et al., 2016; Shukla et al., 2018; Jeon and Kim, 2019; Li et al., 2019). The criteria applied are (1) ‘normal SSR’ as a conventional definition from monoSSR to hexaSSR, (2) ‘extented SSR’ referring from heptaSSR (repeats of 7 bp unit) to decaSSR (repeats of 10 bp unit), and (3) ‘potential SSR’ referring specific cases with only 2 units in pentaSSR and hexaSSR. These criteria have been applied and provided better understanding of SSR patterns in previous analyses in chloroplast genomes of Dysphania species ( Kim et al., 2019f), Arabidopsis thaliana ( Park et al., 2020c), Chenopodium album ( Park et al., 2021b), Diarthron linifolium ( Kim et al., 2021b), and mitochondrial genome of Rosa rugosa ( Park et al., 2020d).
Phylogenetic analysis
Twenty-nine representative Bignoniaceae chloroplast genomes including two C. grandiflora chloroplast genomes and one outgroup species, Paulownia tomentosa ( Yi and Kim, 2016), were used for calculating multiple sequence alignments of 60 conserved genes by MAFFT v7.450 ( Katoh and Standley, 2013) for constructing phylogenetic trees. We used MEGA X ( Kumar et al., 2018) to construct maximum likelihood (ML) and neighbor-joining (NJ) and MrBayes v3.2.6 ( Ronquist et al., 2012) to carry out Bayesian inference (BI). A heuristic search was used with nearest-neighbor interchange branch swapping, the Tamura-Nei model, and uniform rates among sites to construct ML and NJ phylogenetic trees with default values for other options. To estimate the node confidences bootstrap analyses with 1,000 and 10,000 bootstrap pseudoreplicates were conducted for ML and NJ trees, respectively. For the BI analysis, the GTR (general time reversible) model with gamma rates was used as a molecular model and Markov-Chain Monte Carlo algorithm was employed for 1,000,000 generations with four chains running simultaneously. To build the consensus tree of BI, we sampled trees every 200 generations after removing 100,000 generations as a ‘burn-in’.
RESULTS AND DISCUSSION
The chloroplast genome of C. grandiflora (GenBank accession number: OM279807) is 154,293 bp (GC ratio: 38.1%) and has four subregions: 85,078 bp of LSC region (36.2%) and 18,577 bp of SSC region (33.0%) regions are separated by 25,319 bp of IR region (43.2%) ( Fig. 1). Its length is shorter than that of the previous chloroplast genome by 10 bp (154,303 bp; GenBank accession number: MW430049). It contains 132 genes (87 protein-coding genes [PCGs], eight ribosomal RNAs [rRNAs], and 37 transfer RNAs [tRNA]); 19 genes (eight PCGs, four rRNAs, and seven tRNAs) are duplicated in IR regions ( Fig. 1). Structural variation between C. grandiflora and T. capensis was identified using Mauve v1.1.3 ( Darling et al., 2004) in LSC region: the region between 48,536 bp and 73,124 bp in C. grandiflora chloroplast genome was inverted against that of T. capensis. This phenomenon also occurred between two Incarvillea chloroplast genomes in the same family ( Ma et al., 2019; Wu et al., 2021), congruent to the previous study ( Chen et al., 2022). It suggests that inversion events in LSC occurred in Bignoniaceae in comparison to the other families, such as Amaranthaceae ( Park et al., 2021b) and Oleaceae ( Park et al., 2019f). Interspecific variations between the two C. grandiflora chloroplast genomes were investigated. In total, one SNP and five INDEL regions (40 bp in total). One SNP was located between trnC and petN. The 20-bp deletion, which is the longest INDEL, was found in 3' end of ycf1, which expanded two more amino acids ( Fig. 2A). Another 15-bp INDEL region was located in the first intron of accD, presenting the three-time repeat in the chloroplast genome assembled in this study while two-time repeat in the previous chloroplast genome ( Fig. 2B). One-bp deletion was found in the intergenic region between trnK and rps16, exhibiting difference of monoSSR ( Fig. 2C). The two INDEL regions were found in 16S rRNA in the IR region ( Fig. 2D), showing that two-time repeat of CAT was destroyed in the chloroplast genome assembled in this study by this 2-bp deletion ( Fig. 2D). Interestingly, all INDEL regions were linked to the repetitive sequences and proportion of INDEL regions related to SSRs (20%) was low. Numbers of intraspecific variations of C. grandiflora are relatively lower than those identified between the samples between Korea and China ( Fig. 2E, Table 1). This result seems to be incongruent to the previous studies that estimated their genetic diversities using the classical methods ( He and Gu, 1990; Wu et al., 1990; Wen and Jansen, 1995). Therefore, additional C. grandiflora chloroplast genomes will be required to evaluate its genetic diversity. SSR has been utilized as useful molecular markers ( Huang et al., 2015; Li et al., 2020a, 2020b). Seventy-two normal SSRs, 418 potential SSRs, and 47 extended SSRs were identified in both C. grandiflora chloroplast genomes ( Table 2). Most of normal SSRs are monoSSRs ( Fig. 3), which is similar to those of the other plant species ( Kim et al., 2019f, 2021b; Park et al., 2020c, 2021b). Nine normal SSRs and 18 extended SSRs (22.68%) were identified in the genic regions of matK, atpA, rpoC2, psbC, psaI, psbB, rpoA, rpl22, ycf2, ycf1, ndhI, ndhG, and ndhD ( Table 2) and 12 normal SSRs and three extended SSRs (12.61%) were found in the intronic regions of rps16, trnS-CGA, atpF, ycf3, trnL-UAA, petD, rps16, and ndhA ( Table 2). Due to low number of intraspecific variations, only one monoSSR (cM0000002) displayed the differences of the number of repeats between the two chloroplast genomes. These SSRs will be useful to develop molecular makers because high genetic diversity of C. grandiflora was estimated in previous studies ( He and Gu, 1990; Wu et al., 1990; Wen and Jansen, 1995). Three phylogenetic trees showed that C. grandiflora was clustered with T. capensis/ Incarvillea with high supportive values ( Fig. 4). In addition, trees presented that tribes covering more than one chloroplast genome, including Tecomeae, Catalpeae, Crescentiina, and Bignonieae, were well clustered with high supportive values ( Fig. 4). It is congruent to previous phylogenetic studies, except for the Catalpeae and Oroxylae clustered in one clade with week bootstrap values ( Olmstead et al., 2009). It may be caused by different coverage of samples between the two studies. Together with additional chloroplast genomes of Bignoniaceae, C. grandiflora chloroplast genome will help to understand evolutionary history of Bignoniacea.
ACKNOWLEDGMENTS
This study was supported by the InfoBoss Research Grant (IBG-0038).
Fig. 1.
Circular map of Campsis grandiflora complete chloroplast genome. The genes located outside of the circle are transcribed clockwise, while those located inside are transcribed counterclockwise. The dark gray plot in the inner circle corresponds to GC content. Large singlecopy (LSC), small single-copy (SSC), and inverted repeat (IR) are indicated with LSC, SSC, and IR (IRA and IRB), respectively. 
Fig. 2.
Intraspecific variations identified from the two Campsis grandiflora chloroplast genomes. A–D. Consensus sequences were displayed with different background colors of each base. Insertions and deletions were presented as orange-colored boxes. Yellow and red arrows indicate protein-coding genes (PCGs) and rRNAs, respectively. Amino acid sequences in PCG were displayed below the nucleotide sequences. Blue arrows and lines indicate repetitive sequences found in insertions and deletions. E. X-axis means the number of single nucleotide polymorphism (SNPs) and Y-axis indicates insertion and deletion (INDEL) coverage (bp). Blue-colored circle indicates the number of intraspecific variations identified in C. grandiflora chloroplast genomes. Orange, gray, and yellow-colored circles presented those identified between Chinese and Korean isolates, Chinese and Chinese isolates, and Korean and Korean isolates, respectively. 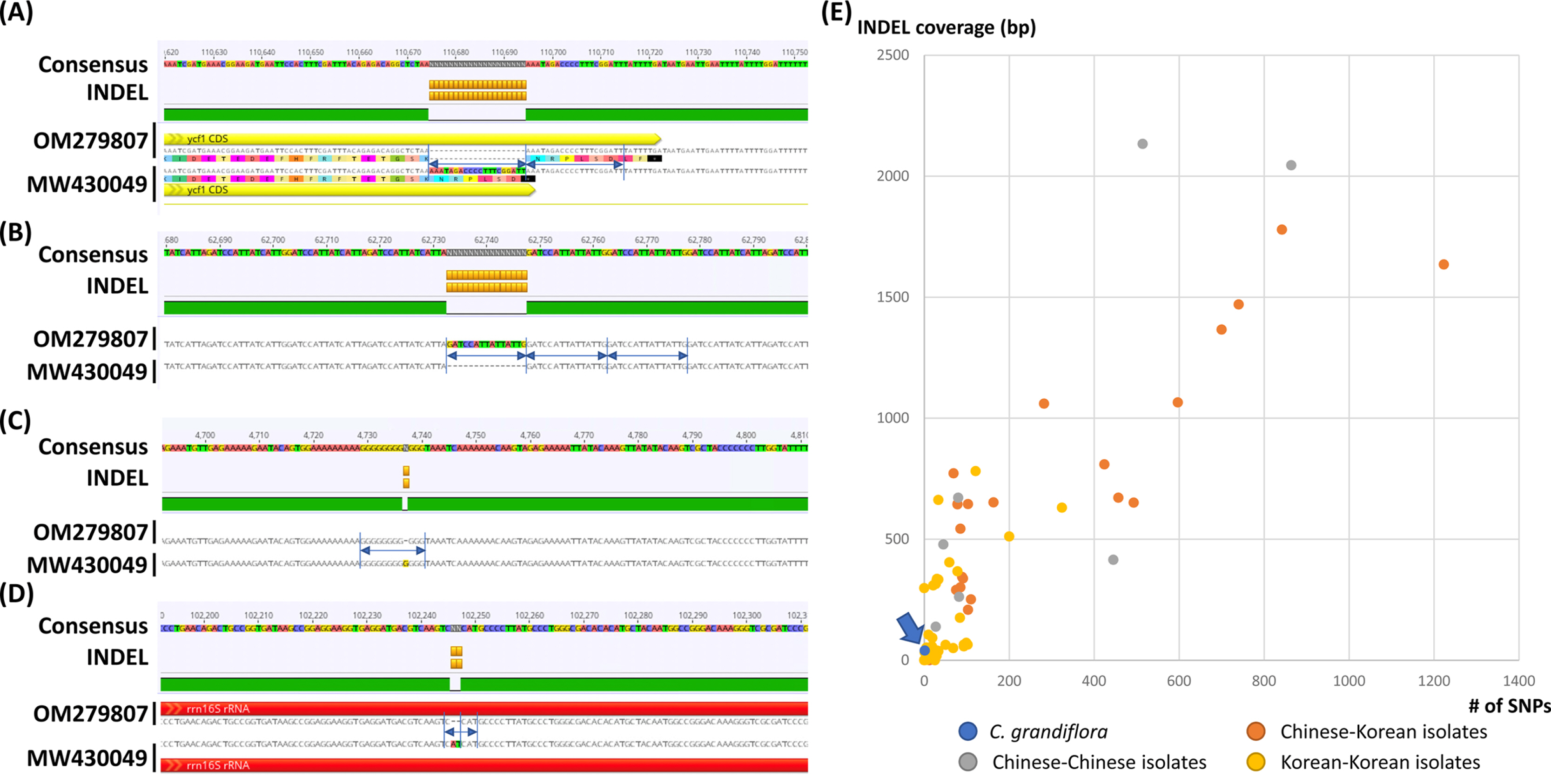
Fig. 3.
Distribution of simple sequence repeats (SSRs) along with types identified from Campsis grandiflora chloroplast genome. X-axis presented SSR types of normal SSRs, potential SSRs, and extended SSRs. Y-axis indicates the number of SSRs. 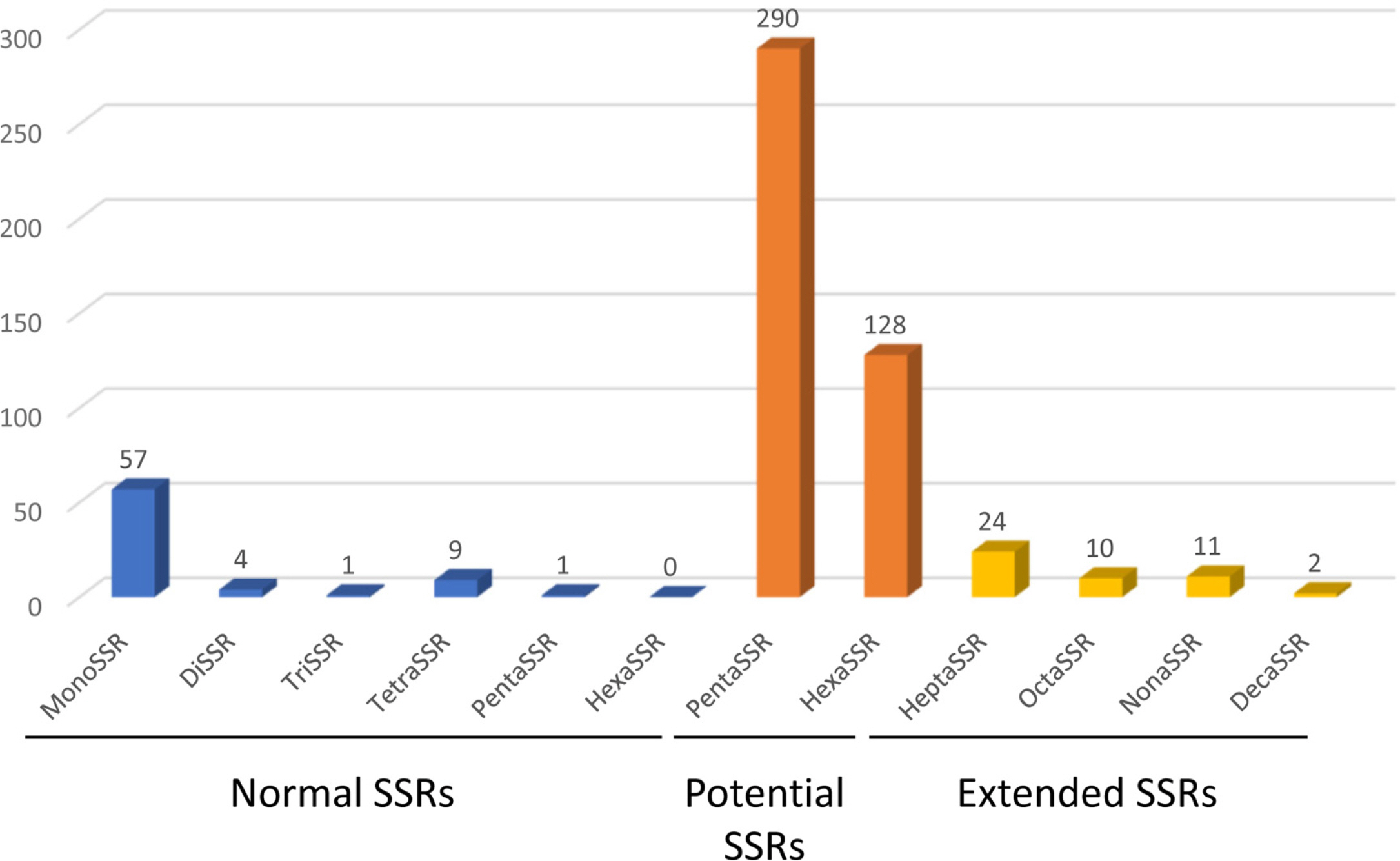
Fig. 4.
Phylogenetic trees of the 29 representative Bignoniaceae chloroplast genomes including two Campsis grandiflora chloroplast genomes. Phylogenetic tree was drawn based on the maximum likelihood phylogenetic tree. The numbers above the branches correspond to the bootstrap support values from the maximum likelihood and neighbor-joining methods, as well as posterior probability from the Bayesian inference. 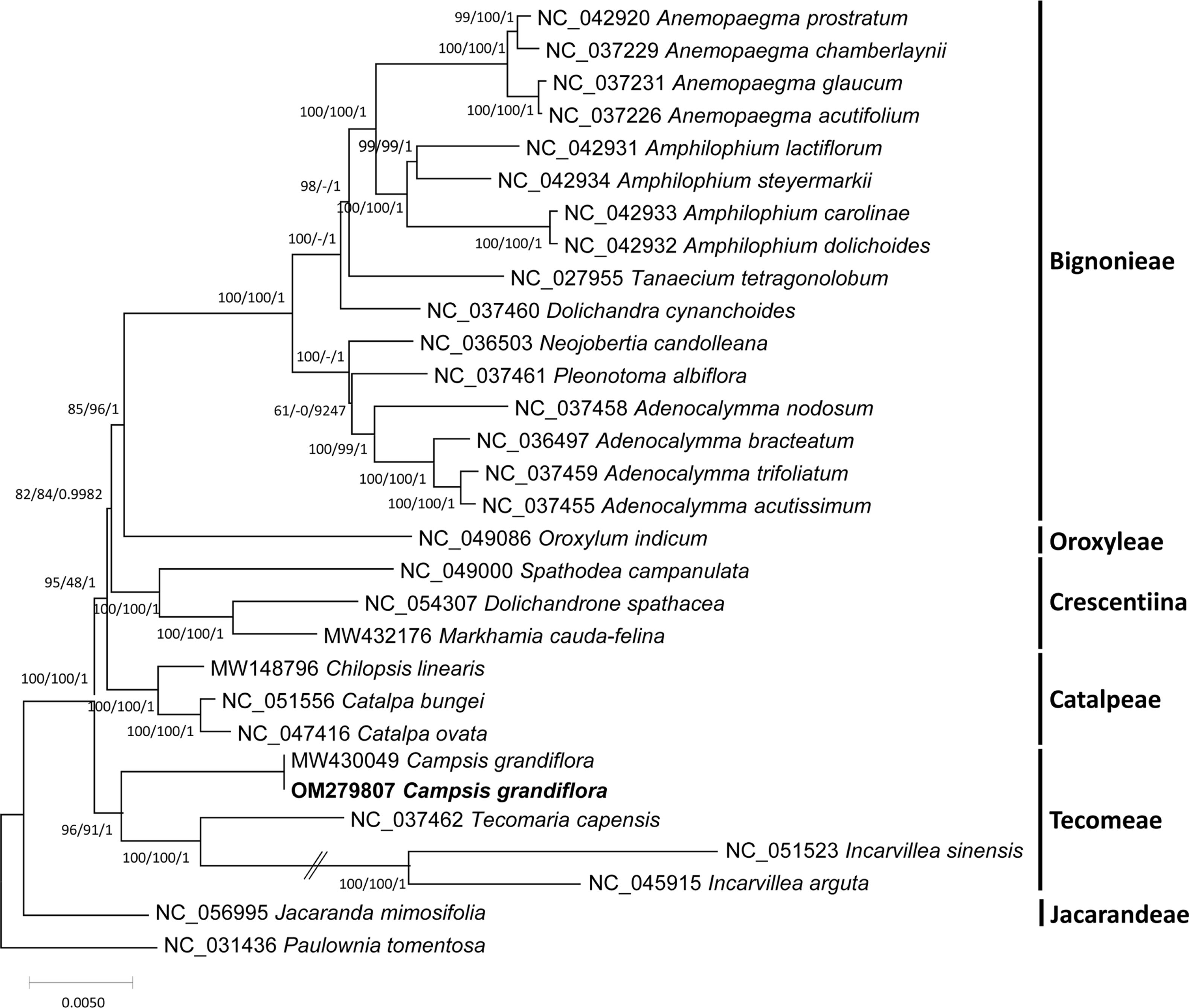
Table 1.
List of numbers of intraspecific variations in the chloroplast genomes identified from the samples isolated from Korea and China.
|
Family |
Species name |
Source |
Target |
No. of SNPs |
INDEL length (bp) |
Reference |
|
Selaginellaceae |
Selaginella tamariscina
|
Korean isolate (MN894555) |
Chinese isolate (NC_041646) |
1,223 |
1,635 |
Park et al. (2020a), Xu et al. (2018)
|
|
Rosaceae |
Hagenia abyssinica
|
Chinese isolate (KX008604) |
Chinese isolate (KY420026) |
82 |
262 |
Gichira et al. (2017), Zhang et al. (2017)
|
|
Sanguisorba officinalis
|
Chinese isolate (NC_044694) |
Korean isolate (MZ145058) |
85 |
301 |
|
|
Chinese isolate (NC_044694) |
Korean isolate (MK696193) |
75 |
290 |
Meng et al. (2018)
|
|
Korean isolate (MZ145058) |
Korean isolate (MK696193) |
10 |
105 |
|
|
Sanguisorba tenuifolia
|
Chinese isolate (NC_044692) |
Chinese isolate (NC_042223) |
27 |
138 |
|
|
Chinese isolate (NC_044692) |
Korean isolate (MK696194) |
91 |
338 |
Meng et al. (2018), Park et al. (2018)
|
|
Chinese isolate (NC_042223) |
Korean isolate (MK696194) |
89 |
344 |
|
|
Sanguisorba stipulata
|
Korean isolate (MZ145059) |
Korean isolate (MK696195) |
78 |
367 |
|
|
Sanguisorba filiformis
|
Chinese isolate (NC_044693) |
Chinese isolate (KY419920) |
45 |
478 |
Meng et al. (2018), Zhang et al. (2017)
|
|
Amaranthaceae |
Chenopodium album
|
Korean isolate (MW446246) |
Korean isolate (MW446245) |
28 |
19 |
Hong et al. (2017), Park et al. (2021b)
|
|
Korean isolate (MW446246) |
Korean isolate (MW446243) |
2 |
2 |
|
|
Korean isolate (MW446246) |
Korean isolate (MW446241) |
27 |
315 |
|
|
Korean isolate (MW446246) |
Korean isolate (MW446242) |
21 |
18 |
|
|
Korean isolate (MW446246) |
Korean isolate (MW446244) |
14 |
33 |
|
|
Korean isolate (MW446246) |
Korean isolate (NC_034950) |
17 |
36 |
|
|
Korean isolate (MW446245) |
Korean isolate (MW446243) |
30 |
35 |
|
|
Korean isolate (MW446245) |
Korean isolate (MW446241) |
0 |
298 |
|
|
Korean isolate (MW446245) |
Korean isolate (MW446242) |
21 |
15 |
|
|
Korean isolate (MW446245) |
Korean isolate (MW446244) |
30 |
40 |
|
|
Korean isolate (MW446245) |
Korean isolate (NC_034950) |
33 |
37 |
|
|
Korean isolate (MW446243) |
Korean isolate (MW446241) |
29 |
331 |
|
|
Korean isolate (MW446243) |
Korean isolate (MW446242) |
23 |
34 |
|
|
Korean isolate (MW446243) |
Korean isolate (MW446244) |
16 |
49 |
|
|
Korean isolate (MW446243) |
Korean isolate (NC_034950) |
19 |
52 |
|
|
Korean isolate (MW446241) |
Korean isolate (MW446242) |
21 |
309 |
|
|
Korean isolate (MW446241) |
Korean isolate (MW446244) |
30 |
336 |
|
|
Korean isolate (MW446241) |
Korean isolate (NC_034950) |
33 |
333 |
|
|
Korean isolate (MW446242) |
Korean isolate (MW446244) |
23 |
39 |
|
|
Korean isolate (MW446242) |
Korean isolate (NC_034950) |
26 |
36 |
|
|
Korean isolate (MW446244) |
Korean isolate (NC_034950) |
18 |
57 |
|
|
Dysphania pumilio
|
Korean isolate (MH936550) |
Korean isolate (MK541016) |
25 |
2 |
Kim et al. (2019b), Park and Kim (2019)
|
|
Suaeda japonica
|
Korean isolate (MK764271) |
Korean isolate (MK558824) |
3 |
3 |
Kim et al. (2019g), Kang et al. (2020)
|
|
Magnoliaceae |
Magnolia kobus
|
Korean isolate (NC_023237) |
Korean isolate (MN894553) |
50 |
63 |
|
|
Liriodendron tulipifera
|
Korean isolate (MK477550) |
Chinese isolate (NC_008326) |
12 |
0 |
Cai et al. (2006), Kwon et al. (2019b)
|
|
Theaceae |
Camellia japonica
|
Korean isolate (MK353210) |
Korean isolate (MK353211) |
25 |
2 |
Kim et al. (2017), Kwon et al. (2019a), Li et al. (2019), Min et al. (2019b)
|
|
Korean isolate (MK353210) |
Korean isolate (KU951523) |
25 |
2 |
|
Korean isolate (MK353211) |
Korean isolate (KU951523) |
25 |
0 |
|
Chinese isolate (MW602996) |
Korean isolate (MK353210) |
8 |
36 |
|
Chinese isolate (MW602996) |
Korean isolate (MK353211) |
8 |
38 |
|
Chinese isolate (MW602996) |
Korean isolate (KU951523) |
33 |
38 |
|
Chinese isolate (NC_036830) |
Korean isolate (MK353210) |
78 |
644 |
|
Chinese isolate (NC_036830) |
Korean isolate (MK353211) |
78 |
645 |
|
Chinese isolate (NC_036830) |
Korean isolate (KU951523) |
103 |
645 |
|
Chinese isolate (NC_036830) |
Chinese isolate (MW602996) |
80 |
671 |
|
Fagaceae |
Fagus multinervis
|
Korean isolate (OM373199) |
Korean isolate (MZ962344) |
4 |
1 |
Park and Oh (2020), Park et al. (2019a), Yang et al. (2020), Park et al. (unpubl. data) |
|
Korean isolate (OM373199) |
Korean isolate (MK518070) |
1 |
2 |
|
Korean isolate (OM373199) |
Korean isolate (MN894556) |
1 |
0 |
|
Korean isolate (OM373199) |
Korean isolate (MT762296) |
1 |
2 |
|
Korean isolate (MZ962344) |
Korean isolate (MK518070) |
3 |
3 |
|
Korean isolate (MZ962344) |
Korean isolate (MN894556) |
5 |
1 |
|
Korean isolate (MZ962344) |
Korean isolate (MT762296) |
3 |
3 |
|
Korean isolate (MK518070) |
Korean isolate (MT762296) |
0 |
0 |
|
Korean isolate (MK518070) |
Korean isolate (MN894556) |
2 |
2 |
|
Korean isolate (MN894556) |
Korean isolate (MT762296) |
2 |
2 |
|
Fagus japonica
|
Korean isolate (NC_053352) |
Korean isolate (MT762295) |
1 |
32 |
Yang et al. (2020), Park et al. (unpubl. data) |
|
Caryophyllaceae |
Pseudostellaria palibiniana
|
Korean isolate (MK120981) |
Korean isolate (MK309611) |
84 |
175 |
Kim et al. (2019c, 2019d)
|
|
Salicaceae |
Salix koriyanagi
|
Korean isolate (MK541017) |
Korean isolate (MK120982) |
0 |
0 |
Kim et al. (2019a), Park et al. (2019d)
|
|
Rosaceae |
Pyrus ussuriensis
|
Korean isolate (MK507863) |
Korean isolate (MK172841) |
121 |
781 |
Cho et al. (2019), Gil et al. (2019)
|
|
Asteraceae |
Artemisia fukudo
|
Korean isolate (NC_044156) |
Korean isolate (MG951488) |
7 |
12 |
Lee et al. (2016), Min et al. (2019c), |
|
Erigeron canadensis
|
Korean isolate (MT806101) |
Chinese isolate (NC_046789) |
103 |
208 |
Park et al. (unpubl. data), Zhang et al. (2019)
|
|
Chrysanthemum zawadskii
|
Korean isolate (MW539687) |
Chinese isolate (MG799556) |
110 |
251 |
Baek et al. (2021), Hongmei et al. (2021)
|
|
Orchidaceae |
Goodyera schlechtendaliana
|
Korean isolate (MK144665) |
Korean isolate (MK134679) |
200 |
511 |
Niu et al. (2017), Oh et al. (2019a, 2019b)
|
|
Chinese isolate (NC_029364) |
Korean isolate (MK144665) |
842 |
1,779 |
|
Chinese isolate (NC_029364) |
Korean isolate (MK134679) |
740 |
1,470 |
|
Chinese isolate (LC085346) |
Chinese isolate (NC_029364) |
514 |
2,133 |
|
Chinese isolate (LC085346) |
Korean isolate (MK144665) |
700 |
1,366 |
|
Chinese isolate (LC085346) |
Korean isolate (MK134679) |
597 |
1,065 |
|
Chinese isolate |
Chinese isolate |
445 |
415 |
|
(AB893949) |
(LC085346) |
|
|
|
Chinese isolate (AB893949) |
Chinese isolate (NC_029364) |
864 |
2,045 |
|
Chinese isolate (AB893949) |
Korean isolate (MK144665) |
282 |
1,060 |
|
Chinese isolate |
Korean isolate |
163 |
652 |
|
(AB893949) |
(MK134679) |
|
|
|
Gastrodia elata
|
Korean isolate (MN026874) |
Korean isolate (MN296709) |
324 |
630 |
Kang et al. (2020), Park et al. (2020b), Yuan et al. (2018)
|
|
Chinese isolate (NC_037409) |
Korean isolate (MN026874) |
493 |
651 |
|
Chinese isolate (NC_037409) |
Korean isolate (MN296709) |
457 |
671 |
|
Oleaceae |
Abeliophyllum distichum
|
Korean isolate (NC_031445) |
Korean isolate (MN127986) |
93 |
57 |
Min et al. (2019a), Park et al. (2019e, 2019f, 2021c)
|
|
Korean isolate (NC_031445) |
Korean isolate (MK616470) |
93 |
64 |
|
Korean isolate (NC_031445) |
Korean isolate (MF407183) |
93 |
57 |
|
Korean isolate (NC_031445) |
Korean isolate (MN116559) |
102 |
64 |
|
Korean isolate (NC_031445) |
Korean isolate (MW426545) |
99 |
72 |
|
Korean isolate (MN127986) |
Korean isolate (MK616470) |
0 |
0 |
|
Korean isolate (MN127986) |
Korean isolate (MF407183) |
0 |
1 |
|
Korean isolate (MN127986) |
Korean isolate (MN116559) |
9 |
12 |
|
Oleaceae |
Abeliophyllum distichum
|
Korean isolate (MN127986) |
Korean isolate (MW426545) |
6 |
20 |
Min et al. (2019a), Park et al. (2019e, 2019f, 2021c)
|
|
Korean isolate (MK616470) |
Korean isolate (MF407183) |
0 |
1 |
|
Korean isolate (MK616470) |
Korean isolate (MN116559) |
9 |
11 |
|
Korean isolate (MK616470) |
Korean isolate (MW426545) |
6 |
19 |
|
Korean isolate (MF407183) |
Korean isolate (MN116559) |
9 |
11 |
|
Korean isolate (MF407183) |
Korean isolate (MW426545) |
6 |
21 |
|
Korean isolate (MN116559) |
Korean isolate (MW426545) |
7 |
23 |
|
Adoxaceae |
Viburnum erosum
|
Korean isolate (MN641480) |
Korean isolate (MN218778) |
16 |
50 |
Choi et al. (2020), Park et al. (2019b)
|
|
Brasicaceae |
Arabidopsis thaliana
|
Korean isolate (MK353213) |
Chinese isolate (MK380719) |
10 |
33 |
Park et al. (2020c)
|
|
Ranunculaceae |
Aconitum coreanum
|
Korean isolate (NC_031421) |
Korean isolate (KU318669) |
29 |
61 |
Kim et al. (2019h), Park et al. (2017)
|
|
Korean isolate (NC_031421) |
Korean isolate (MN400660) |
5 |
52 |
|
Korean isolate (MN400660) |
Korean isolate (KU318669) |
19 |
92 |
|
Thymelaeaceae |
Daphne genkwa
|
Korean isolate (MT754180) |
Korean isolate (Unpub) |
59 |
404 |
Yoo et al. (2021)
|
|
Korean isolate (MT754180) |
Chinese isolate (NC_045891) |
69 |
772 |
|
Korean isolate (Unpub) |
Chinese isolate (NC_045891) |
85 |
543 |
|
Campanulaceae |
Campanula takesimana
|
Korean isolate (MW013763) |
Korean isolate (NC_026203) |
33 |
662 |
Cheon et al. (2016), Park et al. (2021a)
|
|
Poaceae |
Zoysia japonica
|
Korean isolate (MW690657) |
Korean isolate (NC_036827) |
68 |
50 |
Lee and Park (2021)
|
|
Zoysia macrostachya
|
Korean isolate (MZ233426) |
Korean isolate (NC_042189) |
29 |
18 |
Cheon et al. (2021), Oh et al. (2021)
|
|
Staphyleaceae |
Euscaphis japonica
|
Chinese isolate (MN159078) |
Korean isolate (NC_052922) |
424 |
809 |
Oh and Park (2020), Xiang et al. (2019)
|
Table 2.
List of normal SSRs and extended SSRs identified in the C. grandiflora chloroplast genomes isolated in Korea.
|
No. |
Name |
SSRType |
Type |
Start |
End |
Unit sequence |
Repeat No. |
Gene |
Position |
|
1 |
cO0000001 |
ExtendedSSR |
OctaSSR |
2551 |
2566 |
ATAATTGG |
2 |
matK
|
Genic |
|
2 |
cM0000001 |
SSR |
MonoSSR |
4417 |
4427 |
A |
11 |
- |
- |
|
3 |
cM0000002 |
SSR |
MonoSSR |
4729 |
4739 |
G |
11 |
- |
- |
|
4 |
cM0000003 |
SSR |
MonoSSR |
4807 |
4817 |
T |
11 |
- |
- |
|
5 |
cM0000004 |
SSR |
MonoSSR |
5226 |
5235 |
T |
10 |
rps16
|
Intronic |
|
6 |
cM0000005 |
SSR |
MonoSSR |
5249 |
5258 |
A |
10 |
rps16
|
Intronic |
|
7 |
cO0000002 |
ExtendedSSR |
OctaSSR |
6472 |
6487 |
AAATAGAT |
2 |
- |
- |
|
8 |
cM0000006 |
SSR |
MonoSSR |
6513 |
6522 |
T |
10 |
- |
- |
|
9 |
cD0000001 |
SSR |
DiSSR |
7299 |
7310 |
AT |
6 |
- |
- |
|
10 |
cM0000007 |
SSR |
MonoSSR |
8095 |
8105 |
T |
11 |
- |
- |
|
11 |
cHe0000001 |
ExtendedSSR |
HeptaSSR |
8479 |
8492 |
AATGTAA |
2 |
- |
- |
|
12 |
cD0000002 |
SSR |
DiSSR |
8530 |
8539 |
TA |
5 |
- |
- |
|
13 |
cM0000008 |
SSR |
MonoSSR |
8698 |
8709 |
T |
12 |
- |
- |
|
14 |
cM0000009 |
SSR |
MonoSSR |
9087 |
9096 |
A |
10 |
trnS-CGA |
Intronic |
|
15 |
cM0000010 |
SSR |
MonoSSR |
9287 |
9298 |
A |
12 |
trnS-CGA |
Intronic |
|
16 |
cO0000003 |
ExtendedSSR |
OctaSSR |
10230 |
10245 |
TACGTAAG |
2 |
atpA
|
Genic |
|
17 |
cTe0000001 |
SSR |
TetraSSR |
11104 |
11115 |
GTCT |
3 |
atpA
|
Genic |
|
18 |
cM0000011 |
SSR |
MonoSSR |
12359 |
12371 |
T |
13 |
atpF
|
Intronic |
|
19 |
cN0000001 |
ExtendedSSR |
NonaSSR |
13024 |
13041 |
TCTTTTTTA |
2 |
- |
- |
|
20 |
cM0000012 |
SSR |
MonoSSR |
13054 |
13063 |
T |
10 |
- |
- |
|
21 |
cHe0000003 |
ExtendedSSR |
HeptaSSR |
14205 |
14218 |
ATTTATT |
2 |
- |
- |
|
22 |
cHe0000004 |
ExtendedSSR |
HeptaSSR |
14480 |
14493 |
ATTTTTT |
2 |
- |
- |
|
23 |
cM0000013 |
SSR |
MonoSSR |
16315 |
16327 |
T |
13 |
- |
- |
|
24 |
cP0000031 |
SSR |
PentaSSR |
16455 |
16469 |
CAAAT |
3 |
- |
- |
|
25 |
cHe0000005 |
ExtendedSSR |
HeptaSSR |
17159 |
17172 |
CAACCCT |
2 |
rpoC2
|
Genic |
|
26 |
cM0000014 |
SSR |
MonoSSR |
18522 |
18532 |
T |
11 |
rpoC2
|
Genic |
|
27 |
cD0000003 |
SSR |
DiSSR |
19907 |
19916 |
AT |
5 |
rpoC2
|
Genic |
|
28 |
cHe0000006 |
ExtendedSSR |
HeptaSSR |
27420 |
27433 |
TGTATAA |
2 |
- |
- |
|
29 |
cHe0000007 |
ExtendedSSR |
HeptaSSR |
27459 |
27472 |
TGTATAA |
2 |
- |
- |
|
30 |
cM0000015 |
SSR |
MonoSSR |
30555 |
30567 |
A |
13 |
- |
- |
|
31 |
cHe0000008 |
ExtendedSSR |
HeptaSSR |
30733 |
30746 |
AAAGAAA |
2 |
- |
- |
|
32 |
cHe0000009 |
ExtendedSSR |
HeptaSSR |
30834 |
30847 |
TATTGGA |
2 |
- |
- |
|
33 |
cT0000001 |
SSR |
TriSSR |
35063 |
35074 |
TTC |
4 |
psbC
|
Genic |
|
34 |
cM0000016 |
SSR |
MonoSSR |
35277 |
35291 |
T |
15 |
- |
- |
|
35 |
cO0000004 |
ExtendedSSR |
OctaSSR |
35310 |
35325 |
TCGATTTT |
2 |
- |
- |
|
36 |
cM0000017 |
SSR |
MonoSSR |
35664 |
35673 |
A |
10 |
- |
- |
|
37 |
cM0000018 |
SSR |
MonoSSR |
36062 |
36072 |
C |
11 |
- |
- |
|
38 |
cM0000019 |
SSR |
MonoSSR |
36073 |
36084 |
A |
12 |
- |
- |
|
39 |
cM0000020 |
SSR |
MonoSSR |
36178 |
36187 |
A |
10 |
- |
- |
|
40 |
cHe0000011 |
ExtendedSSR |
HeptaSSR |
42112 |
42125 |
TTAATAT |
2 |
- |
- |
|
41 |
cM0000021 |
SSR |
MonoSSR |
42252 |
42261 |
A |
10 |
- |
- |
|
42 |
cTe0000002 |
SSR |
TetraSSR |
42470 |
42481 |
TAAA |
3 |
- |
- |
|
43 |
cM0000022 |
SSR |
MonoSSR |
43350 |
43361 |
T |
12 |
ycf3
|
Intronic |
|
44 |
cO0000005 |
ExtendedSSR |
OctaSSR |
44658 |
44673 |
ATCCTAAT |
2 |
- |
- |
|
45 |
cM0000023 |
SSR |
MonoSSR |
45132 |
45146 |
A |
15 |
- |
- |
|
46 |
cM0000024 |
SSR |
MonoSSR |
46347 |
46359 |
T |
13 |
- |
- |
|
47 |
cD0000004 |
SSR |
DiSSR |
46520 |
46533 |
TA |
7 |
- |
- |
|
48 |
cHe0000012 |
ExtendedSSR |
HeptaSSR |
46676 |
46689 |
AAAAAAT |
2 |
- |
- |
|
49 |
cM0000025 |
SSR |
MonoSSR |
46996 |
47007 |
T |
12 |
- |
- |
|
50 |
cHe0000013 |
ExtendedSSR |
HeptaSSR |
47052 |
47065 |
TATATTT |
2 |
- |
- |
|
51 |
cM0000026 |
SSR |
MonoSSR |
47199 |
47212 |
A |
14 |
- |
- |
|
52 |
cHe0000014 |
ExtendedSSR |
HeptaSSR |
47362 |
47375 |
TCCTATA |
2 |
- |
- |
|
53 |
cM0000027 |
SSR |
MonoSSR |
47393 |
47402 |
T |
10 |
- |
- |
|
54 |
cM0000028 |
SSR |
MonoSSR |
47671 |
47681 |
A |
11 |
trnL-UAA |
Intronic |
|
55 |
cHe0000015 |
ExtendedSSR |
HeptaSSR |
47801 |
47814 |
ATATCAA |
2 |
trnL-UAA |
Intronic |
|
56 |
cHe0000016 |
ExtendedSSR |
HeptaSSR |
48224 |
48237 |
AATTAAG |
2 |
- |
- |
|
57 |
cN0000006 |
ExtendedSSR |
NonaSSR |
48587 |
48604 |
ATGATAAAG |
2 |
- |
- |
|
58 |
cN0000007 |
ExtendedSSR |
NonaSSR |
48643 |
48660 |
AAAGTGAAT |
2 |
- |
- |
|
59 |
cM0000029 |
SSR |
MonoSSR |
50718 |
50730 |
A |
13 |
- |
- |
|
60 |
cHe0000017 |
ExtendedSSR |
HeptaSSR |
51214 |
51227 |
ATTAGTT |
2 |
- |
- |
|
61 |
cDe0000004 |
ExtendedSSR |
DecaSSR |
51678 |
51697 |
TATGAGAAAA |
2 |
- |
- |
|
62 |
cM0000030 |
SSR |
MonoSSR |
51739 |
51748 |
T |
10 |
- |
- |
|
63 |
cTe0000003 |
SSR |
TetraSSR |
52184 |
52195 |
GTTT |
3 |
- |
- |
|
64 |
cO0000006 |
ExtendedSSR |
OctaSSR |
53336 |
53351 |
ATATATAA |
2 |
- |
- |
|
65 |
cM0000031 |
SSR |
MonoSSR |
54063 |
54073 |
A |
11 |
- |
- |
|
66 |
cM0000032 |
SSR |
MonoSSR |
56504 |
56514 |
A |
11 |
- |
- |
|
67 |
cO0000007 |
ExtendedSSR |
OctaSSR |
59209 |
59224 |
TATCAAAA |
2 |
- |
- |
|
68 |
cM0000033 |
SSR |
MonoSSR |
59479 |
59489 |
A |
11 |
- |
- |
|
69 |
cTe0000004 |
SSR |
TetraSSR |
59496 |
59507 |
TTTC |
3 |
- |
- |
|
70 |
cM0000034 |
SSR |
MonoSSR |
59697 |
59709 |
A |
13 |
- |
- |
|
71 |
cO0000008 |
ExtendedSSR |
OctaSSR |
60779 |
60794 |
ATAAAGAA |
2 |
psaI
|
Genic |
|
72 |
cTe0000005 |
SSR |
TetraSSR |
63789 |
63800 |
TTTA |
3 |
|
|
|
73 |
cM0000035 |
SSR |
MonoSSR |
66657 |
66666 |
A |
10 |
atpB
|
Genic |
|
74 |
cDe0000005 |
ExtendedSSR |
DecaSSR |
68648 |
68667 |
AAAATCAATA |
2 |
- |
- |
|
75 |
cM0000036 |
SSR |
MonoSSR |
70334 |
70344 |
A |
11 |
- |
- |
|
76 |
cM0000037 |
SSR |
MonoSSR |
70396 |
70405 |
A |
10 |
- |
- |
|
77 |
cM0000038 |
SSR |
MonoSSR |
72938 |
72949 |
A |
12 |
- |
- |
|
78 |
cHe0000021 |
ExtendedSSR |
HeptaSSR |
72980 |
72993 |
ATTTAAG |
2 |
- |
- |
|
79 |
cTe0000006 |
SSR |
TetraSSR |
73132 |
73143 |
AAAG |
3 |
- |
- |
|
80 |
cHe0000022 |
ExtendedSSR |
HeptaSSR |
73526 |
73539 |
CTGGTTG |
2 |
psbB
|
Genic |
|
81 |
cHe0000023 |
ExtendedSSR |
HeptaSSR |
73990 |
74003 |
GGCGTGG |
2 |
psbB
|
Genic |
|
82 |
cM0000039 |
SSR |
MonoSSR |
75085 |
75094 |
C |
10 |
- |
- |
|
83 |
cM0000040 |
SSR |
MonoSSR |
75096 |
75108 |
A |
13 |
- |
- |
|
84 |
cM0000041 |
SSR |
MonoSSR |
76129 |
76140 |
A |
12 |
- |
- |
|
85 |
cO0000011 |
ExtendedSSR |
OctaSSR |
77589 |
77604 |
ATACAGAA |
2 |
petD
|
Intronic |
|
86 |
cM0000042 |
SSR |
MonoSSR |
79112 |
79125 |
T |
14 |
rpoA
|
Genic |
|
87 |
cN0000009 |
ExtendedSSR |
NonaSSR |
79940 |
79957 |
TTTCTCTTG |
2 |
- |
- |
|
88 |
cM0000043 |
SSR |
MonoSSR |
81025 |
81035 |
T |
11 |
- |
- |
|
89 |
cM0000044 |
SSR |
MonoSSR |
81538 |
81550 |
T |
13 |
- |
- |
|
90 |
cM0000045 |
SSR |
MonoSSR |
82710 |
82725 |
T |
16 |
rpl16
|
Intronic |
|
91 |
cM0000046 |
SSR |
MonoSSR |
82788 |
82800 |
A |
13 |
rpl16
|
Intronic |
|
92 |
cO0000012 |
ExtendedSSR |
OctaSSR |
83219 |
83234 |
TTTCTCTC |
2 |
rpl16
|
Intronic |
|
93 |
cHe0000026 |
ExtendedSSR |
HeptaSSR |
84484 |
84497 |
TTACTAA |
2 |
rpl22
|
Genic |
|
94 |
cM0000047 |
SSR |
MonoSSR |
85098 |
85107 |
T |
10 |
- |
- |
|
95 |
cN0000010 |
ExtendedSSR |
NonaSSR |
89648 |
89665 |
GGAACATTT |
2 |
ycf2
|
Genic |
|
96 |
cN0000011 |
ExtendedSSR |
NonaSSR |
92241 |
92258 |
GATATTGAT |
2 |
ycf2
|
Genic |
|
97 |
cN0000012 |
ExtendedSSR |
NonaSSR |
92285 |
92302 |
TATTGATGC |
2 |
ycf2
|
Genic |
|
98 |
cHe0000027 |
ExtendedSSR |
HeptaSSR |
92995 |
93008 |
ACTTGGA |
2 |
ycf2
|
Genic |
|
99 |
cM0000048 |
SSR |
MonoSSR |
112535 |
112544 |
A |
10 |
ycf1
|
Genic |
|
100 |
cM0000049 |
SSR |
MonoSSR |
113091 |
113102 |
A |
12 |
ycf1
|
Genic |
|
101 |
cTe0000007 |
SSR |
TetraSSR |
115405 |
115416 |
GATT |
3 |
- |
- |
|
102 |
cM0000050 |
SSR |
MonoSSR |
118228 |
118239 |
T |
12 |
ndhA
|
Intronic |
|
103 |
cM0000051 |
SSR |
MonoSSR |
118287 |
118296 |
A |
10 |
ndhA
|
Intronic |
|
104 |
cM0000052 |
SSR |
MonoSSR |
118446 |
118455 |
T |
10 |
ndhA
|
Intronic |
|
105 |
cHe0000030 |
ExtendedSSR |
HeptaSSR |
119608 |
119621 |
ATTGGAA |
2 |
ndhI
|
Genic |
|
106 |
cTe0000008 |
SSR |
TetraSSR |
120108 |
120119 |
TTCA |
3 |
- |
- |
|
107 |
cHe0000031 |
ExtendedSSR |
HeptaSSR |
120362 |
120375 |
TATTCTA |
2 |
ndhG
|
Genic |
|
108 |
cM0000053 |
SSR |
MonoSSR |
121803 |
121816 |
T |
14 |
- |
- |
|
109 |
cN0000013 |
ExtendedSSR |
NonaSSR |
122187 |
122204 |
CTACTTTAG |
2 |
ndhD
|
Genic |
|
110 |
cTe0000009 |
SSR |
TetraSSR |
123187 |
123198 |
TATT |
3 |
ndhD
|
Genic |
|
111 |
cM0000054 |
SSR |
MonoSSR |
125479 |
125488 |
T |
10 |
- |
- |
|
112 |
cM0000055 |
SSR |
MonoSSR |
125903 |
125915 |
A |
13 |
- |
- |
|
113 |
cHe0000032 |
ExtendedSSR |
HeptaSSR |
126066 |
126079 |
TTTTTTA |
2 |
- |
- |
|
114 |
cM0000056 |
SSR |
MonoSSR |
128639 |
128648 |
A |
10 |
- |
- |
|
115 |
cHe0000034 |
ExtendedSSR |
HeptaSSR |
146362 |
146375 |
GTTCCAA |
2 |
ycf2
|
Genic |
|
116 |
cN0000014 |
ExtendedSSR |
NonaSSR |
147070 |
147087 |
GCATCAATA |
2 |
ycf2
|
Genic |
|
117 |
cN0000015 |
ExtendedSSR |
NonaSSR |
147110 |
147127 |
TATCATCAA |
2 |
ycf2
|
Genic |
|
118 |
cN0000016 |
ExtendedSSR |
NonaSSR |
149707 |
149724 |
AAATGTTCC |
2 |
ycf2
|
Genic |
|
119 |
cM0000057 |
SSR |
MonoSSR |
154265 |
154274 |
A |
10 |
|
|
LITERATURE CITED
Baek, J. Park, S. Lee, J. Min, J. Park, J and Lee, GW. 2021. The complete chloroplast genome of Chrysanthemum zawadskii Herbich (Asteraceae) isolated in Korea. Mitochondrial DNA Part B Resources 6: 1956-1958.    Cai, Z. Penaflor, C. Kuehl, JV. Leebens-Mack, J. Carlson, JE. dePamphilis, CW. Boore, JL and Jansen, RK. 2006. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: Implications for the phylogenetic relationships of magnoliids. BMC Evolutionary Biology 6: 77.     Chen, H. Chen, Z. Du, Q. Jiang, M. Wang, B and Liu, C. 2022. Complete chloroplast genome of Campsis grandiflora (Thunb.) schum and systematic and comparative analysis within the family Bignoniaceae. Molecular Biology Reports 49: 3085-3098.    Chen, J.. Hao, Z. Xu, H. Yang, L. Liu, G. Sheng, Y. Zheng, C. Zheng, W. Cheng, T and Shi, J. 2015. The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Hu et Cheng. Frontiers in Plant Science 6: 447.  Cheng, J. Zhao, Z. Li, B. Qin, C. Wu, Z. Trejo-Saavedra, DL. Luo, X. Cui, J. Rivera-Bustamante, RF. Li, S and Hu, K. 2016. A comprehensive characterization of simple sequence repeats in pepper genomes provides valuable resources for marker development in Capsicum
. Scientific Reports 6: 18919.     Cheon, K-S. Kim, K-A. Jang, S-K and Yoo, K-O. 2016. Complete chloroplast genome sequence of Campanula takesimana (Campanulaceae), an endemic to Korea. Mitochondrial DNA Part A DNA Mapping, Sequencing, and Analysis 27: 2169-2171.   Cheon, S-H. Woo, M-A. Jo, S. Kim, Y-K and Kim, K-J. 2021. The chloroplast phylogenomics and systematics of Zoysia (Poaceae). Plants 10: 1517.    Cho, M-S. Kim, Y. Kim, S-C and Park, J. 2019. The complete chloroplast genome of Korean Pyrus ussuriensis Maxim. (Rosaceae): Providing genetic background of two types of P. ussuriensis
. Mitochondrial DNA Part B Resources 4: 2424-2425.     Choi, YG. Yun, N. Park, J. Xi, H. Min, J. Kim, Y and Oh, S-H. 2020. The second complete chloroplast genome sequence of the Viburnum erosum (Adoxaceae) showed a low level of intra-species variations. Mitochondrial DNA Part B Resources 5: 271-272.    Cui, X-Y. Kim, J-H. Zhao, X. Chen, B-Q. Lee, B-C. Pyo, H-B. Yun, Y-P and Zhang, Y-H. 2006. Antioxidative and acute anti-inflammatory effects of Campsis grandiflora flower. Journal of Ethnopharmacology 103: 223-228.   Fonseca, LHM and Lohmann, LG. 2018. Combining high-throughput sequencing and targeted loci data to infer the phylogeny of the “Adenocalymma-Neojobertia” clade (Bignonieae, Bignoniaceae). Molecular Phylogenetics and Evolution 123: 1-15.   Gil, H-Y. Kim, Y. Kim, S-H. Jeon, J-H. Kwon, Y. Kim, S-C and Park, J. 2019. The complete chloroplast genome of Pyrus ussuriensis Maxim. (Rosaceae). Mitochondrial DNA Part B Resources 4: 1000-1001.   Han, XH. Oh, J-H. Hong, SS. Lee, C. Park, JI. Lee, MK. Hwang, BY and Lee, M-S. 2012. Novel iridoids from the flowers of Campsis grandiflora
. Archives of Pharmacal Research 35: 327-332.    He, S and Gu, Y. 1990. A study on mutual introduction of main woody species between China and the United States. Bulletin of the Nanjing Botanical Garden 1990: 1-21.
Hong, S-Y. Cheon, K-S. Yoo, K-O. Lee, H-O. Cho, K-S. Suh, J-T. Kim, S-J. Nam, J-H. Sohn, H-B and Kim, Y-H. 2017. Complete chloroplast genome sequences and comparative analysis of Chenopodium quinoa and C. album
. Frontiers in Plant Science 8: 1696.    Hongmei, S. Wenrui, H. Dianyun, H and Yang, X. 2021. Complete chloroplast genome sequence of Dendranthema zawadskii Herbich. Mitochondrial DNA Part B Resources 6: 2117-2119.    Huang, J. Yang, X. Zhang, C. Yin, X. Liu, S and Li, X. 2015. Development of chloroplast microsatellite markers and analysis of chloroplast diversity in Chinese jujube ( Ziziphus jujuba Mill.) and wild jujube ( Ziziphus acidojujuba Mill.). PLoS One 10: e0134519.    Jeon, J-H and Kim, S-C. 2019. Comparative analysis of the complete chloroplast genome sequences of three closely related East-Asian wild roses ( Rosa sect. Synstylae; Rosaceae). Genes 10: 23.    Jia, J. Jiang, W. Wei, J. Weng, M and Han, J. 2012. Landscape characters of Campsis grandiflora and its development and application. Acta Agriculturae Jiangxi 24: 66-68.
Jin, JL. Lee, S. Lee, Y-Y. Heo, JE. Kim, JM and Yun-Choi, HS. 2005. Two new non-glycosidic iridoids from the leaves of Campsis grandiflora
. Planta Medica 71: 578-580.   Kang, M-J. Kim, S-C. Lee, H-R. Lee, S-A. Lee, J-W. Kim, T-D and Park, E-J. 2020. The complete chloroplast genome of Korean Gastrodia elata Blume. Mitochondrial DNA Part B Resources 5: 1015-1016.  Kim, D-H. Han, K-M. Bang, M-H. Lee, Y-H. Chung, I-S. Kim, D-K. Kim, S-H. Kwon, B-M. Park, M-H and Baek, N-I. 2007. Cyclohexylethanoids from the flower of Campsis grandiflora
. Bulletin of the Korean Chemical Society 28: 1851-1853.  Kim, D-H. Han, K-M. Chung, I-S. Kim, D-K. Kim, S-H. Kwon, B-M. Jeong, T-S. Park, M-H. Ahn, E-M and Baek, N-I. 2005. Triterpenoids from the flower of Campsis grandiflora K. Schum. as human acyl-CoA: Cholesterol acyltransferase inhibitors. Archives of Pharmacal Research 28: 550-556.    Kim, J. Kim, Y and Park, J. 2019a. Complete chloroplast genome sequence of the Salix koriyanagi Kimura ex Goerz (Salicaceae). Mitochondrial DNA Part B Resources 4: 549-550.  Kim, M-H. Park, S. Lee, J. Baek, J. Park, J and Lee, GW. 2021a. The complete chloroplast genome of Glycyrrhiza uralensis Fisch. isolated in Korea (Fabaceae). Korean Journal of Plant Taxonomy 51: 353-362.   Kim, S-T. Oh, S-H and Park, J. 2021b. The complete chloroplast genome of Diarthron linifolium (Thymelaeaceae), a species found on a limestone outcrop in eastern Asia. Korean Journal of Plant Taxonomy 51: 345-352.   Kim, Y. Chung, Y and Park, J. 2019b. The complete chloroplast genome sequence of Dysphania pumilio (R. Br.) Mosyakin and Clemants (Amaranthaceae). Mitochondrial DNA Part B Resources 4: 403-404.
Kim, Y. Chung, Y and Park, J. 2020. The complete chloroplast genome of Oxybasis glauca (L.) S. Fuentes, Uotila and Borsch (Amaranthaceae) as the first chloroplast genome in genus Oxybasis
. Mitochondrial DNA Part B Resources 5: 1410-1412.
Kim, Y. Heo, K-I. Lee, S and Park, J. 2019c. The complete chloroplast genome sequence of Pseudostellaria palibiniana (Takeda) Ohwi (Caryophyllaceae). Mitochondrial DNA Part B Resources 4: 973-974.   Kim, Y. Heo, K-I and Park, J. 2019d. The second complete chloroplast genome sequence of Pseudostellaria palibiniana (Takeda) Ohwi (Caryophyllaceae): Intraspecies variations based on geographical distribution. Mitochondrial DNA Part B Resources 4: 1310-1311.   Kim, Y. Min, J. Kwon, W. Song, MJ. Nam, S and Park, J. 2019e. The complete chloroplast genome sequence of the Nymphaea capensis Thunb. (Nymphaeaceae). Mitochondrial DNA Part B Resources 4: 401-402.  Kim, Y. Park, J and Chung, Y. 2019f. Comparative analysis of chloroplast genome of Dysphania ambrosioides (L.) Mosyakin and Clemants understanding phylogenetic relationship in genus Dysphania R. Br. Korean Journal of Plant Resources 32: 644-668.
Kim, Y. Park, J and Chung, Y. 2019g. The complete chloroplast genome of Suaeda japonica Makino (Amaranthaceae). Mitochondrial DNA Part B Resources 4: 1505-1507.   Kim, Y. Yi, J-S. Min, J. Xi, H. Kim, DY. Son, J. Park, J and Jeon, J-I. 2019h. The complete chloroplast genome of Aconitum coreanum (H. Lév.) Rapaics (Ranunculaceae). Mitochondrial DNA Part B Resources 4: 3404-3406.    Kwon, W. Kim, Y and Park, J. 2019a. The complete mitochondrial genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. and Boisselier: Inverted repeats on mitochondrial genome between Korean and Japanese isolates. Mitochondrial DNA Part B Resources 4: 769-770.
Lee, B and Park, J. 2021. The complete chloroplast genome of Zoysia japonica Steud. isolated in Korea (Poaceae): Investigation of potential molecular markers on Z. japonica chloroplast genomes. Plant Biotechnology Reports 15: 707-715.   Lee, YS. Park, JY. Kim, J.-K. Lee, HO. Park, H-S. Lee, S-C. Kang, JH. Lee, TJ. Sung, SH and Yang, T-J. 2016. Complete chloroplast genome sequence of Artemisia fukudo Makino (Asteraceae). Mitochondrial DNA Part B Resources 1: 376-377.     Li, W. Zhang, C. Guo, X. Liu, Q and Wang, K. 2019. Complete chloroplast genome of Camellia japonica genome structures, comparative and phylogenetic analysis. PLoS ONE 14: e0216645.    Ma, G-T. Yang, J-G. Zhang, Y-F and Guan, T-X. 2019. Characterization of the complete chloroplast genome of Incarvillea arguta (Bignoniaceae). Mitochondrial DNA Part B Resources 4: 1603-1604.   Meng, X-X. Xian, Y-F. Xiang, L. Zhang, D. Shi, Y-H. Wu, M-L. Dong, G-Q. Ip, S-P. Lin, Z-X and Sun, W. 2018. Complete chloroplast genomes from Sanguisorba: Identity and variation among four species. Molecules 23: 2137.    Min, J. Kim, Y. Xi, H. Jang, T. Kim, G. Park, J and Park, J-H. 2019a. The complete chloroplast genome of a new candidate cultivar, Sang Jae, of Abeliophyllum distichum Nakai (Oleaceae): Initial step of A. distichum intraspecies variations atlas. Mitochondrial DNA Part B Resources 4: 3716-3718.    Min, J. Kwon, W. Xi, H and Park, J. 2019b. The complete chloroplast genome of Leucobryum juniperoideum (brid.) C. Müll. (Leucobryaceae, Bryophyta). Mitochondrial DNA Part B Resources 4: 2962-2963.    Min, J. Park, J. Kim, Y and Kwon, W. 2019c. The complete chloroplast genome of Artemisia fukudo Makino (Asteraceae): Providing insight of intraspecies variations. Mitochondrial DNA Part B Resources 4: 1510-1512.   Oh, S-D. Lee, S-K. Yun, D-W. Sun, H-J. Kang, H-G. Lee, H-Y. Xi, H. Park, J and Lee, B. 2021. The complete chloroplast genome of Zoysia macrostachya (Poaceae): Insights into intraspecific variations and species delimitation of the Zoysia species. Korean Journal of Plant Taxonomy 51: 326-331.   Oh, S-H and Park, J. 2020. The complete chloroplast genome of Euscaphis japonica (Thunb.) Kanitz (Staphyleaceae) isolated in Korea. Mitochondrial DNA Part B Resources 5: 3769-3771.  Oh, S-H. Suh, H-J. Park, J. Kim, Y and Kim, S. 2019a. The complete chloroplast genome sequence of a morphotype of Goodyera schlechtendaliana (Orchidaceae) with the column appendages. Mitochondrial DNA Part B 4: 626-627.
Oh, S-H. Suh, HJ. Park, J. Kim, Y and Kim, S. 2019b. The complete chloroplast genome sequence of Goodyera schlechtendaliana in Korea (Orchidaceae). Mitochondrial DNA Part B Resources 4: 2692-2693.    Oku, H. Iwaoka, E. Shinga, M. Yamamoto, E. Iinuma, M and Ishiguro, K. 2019. Effect of the dried flowers of Campsis grandiflora on stagnant blood syndrome. Natural Product Communications 14: 1-5.   Park, I. Kim, W-J. Yang, S. Yeo, S-M. Li, H and Moon, BC. 2017. The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLoS ONE 12: e0184257.    Park, I. Yang, S. Kim, WJ. Noh, P. Lee, HO and Moon, BC. 2018. Complete chloroplast genome of Sanguisorba × tenuifolia Fisch. ex Link. Mitochondrial DNA Part B Resources 3: 909-910.    Park, J-S. Jin, D-P. Park, J-W and Choi, B-H. 2019a. Complete chloroplast genome of Fagus multinervis, a beech species endemic to Ulleung Island in South Korea. Mitochondrial DNA Part B Resources 4: 1698-1699.   Park, J. Bae, Y. Kim, B-Y. Nam, G-H. Park, J-M. Lee, BY. Suh, H-J and Oh, S-H. 2021a. The complete chloroplast genome of Campanula takesimana Nakai from DokdoIsland in Korea (Campanulaceae). Mitochondrial DNA Part B Resources 6: 135-137.    Park, J. Choi, YG. Yun, N. Xi, H. Min, J. Kim, Y and Oh, S-H. 2019b. The complete chloroplast genome sequence of Viburnum erosum (Adoxaceae). Mitochondrial DNA Part B Resources 4: 3278-3279.    Park, J. Heo, K-I. Kim, Y and Kwon, W. 2019c. The complete chloroplast genome of Potentilla fragarioides var. major Maxim. Mitochondrial DNA Part B Resources 4: 1265-1266.   Park, J and Kim, Y. 2019. The second complete chloroplast genome of Dysphania pumilio (R. Br.) mosyakin and clemants (Amranthaceae): Intraspecies variation of invasive weeds. Mitochondrial DNA Part B Resources 4: 1428-1429.
Park, J. Kim, Y. Lee, G-H and Park, C-H. 2020a. The complete chloroplast genome of Selaginella tamariscina (Beauv.) Spring (Selaginellaceae) isolated in Korea. Mitochondrial DNA Part B Resources 5: 1654-1656.  Park, J. Kim, Y and Xi, H. 2019d. The complete chloroplast genome sequence of male individual of Korean endemic willow, Salix koriyanagi Kimura ex Goerz (Salicaceae). Mitochondrial DNA Part B Resources 4: 1619-1621.   Park, J. Kim, Y. Xi, H. Jang, T and Park, J-H. 2019e. The complete chloroplast genome of Abeliophyllum distichum Nakai (Oleaceae), cultivar Ok Hwang 1ho: Insights of cultivar specific variations of A. distichum
. Mitochondrial DNA Part B Resources 4: 1640-1642.   Park, J. Lee, J and Park, J. 2022. The investigation of intraspecific characteristics and comparative analyses of the complete mitochondrial genome of Stegobium paniceum (Linnaeus, 1758) (Coleoptera: Ptinidae) assembled from public NGS raw reads of the black truffle, Tuber melanosporum
. Science Progress 105: 1-31.    Park, J. Min, J. Kim, Y and Chung, Y. 2021b. The comparative analyses of six complete chloroplast genomes of morphologically Ddiverse Chenopodium album L. (Amaranthaceae) collected in Korea. International Journal of Genomics 2021: 6643444.
Park, J. Min, J. Kim, Y. Xi, H. Kwon, W. Jang, T. Kim, G and Park, J- H. 2019f. The complete chloroplast genome of a new candidate cultivar, Dae Ryun, of Abeliophyllum distichum Nakai (Oleaceae). Mitochondrial DNA Part B Resources 4: 3713-3715.    Park, J and Oh, S-H. 2020. A second complete chloroplast genome sequence of Fagus multinervis Nakai (Fagaceae): Intraspecific variations on chloroplast genome. Mitochondrial DNA Part B Resources 5: 1868-1869.  Park, J. Park, S. Jang, T. Kim, G and Park, J-H. 2021c. The complete chloroplast genome of Abeliophyllum distichum f. lilacinum Nakai (Oleaceae) from the Chungbuk Province, Korea. Mitochondrial DNA Part B Resources 6: 1754-1756.    Park, J. Suh, Y and Kim, S. 2020b. A complete chloroplast genome sequence of Gastrodia elata (Orchidaceae) represents high sequence variation in the species. Mitochondrial DNA Part B Resources 5: 517-519.    Park, J and Xi, H. 2021. Investigation of nucleotide diversity based on 17 sea cucumber mitochondrial genomes and assessment of sea cucumber mitochondrial gene markers. Advances in Oceanography and Marine Biology 2: 2021.  Park, J. Xi, H and Kim, Y. 2020c. The complete chloroplast genome of Arabidopsis thaliana isolated in Korea (Brassicaceae): An investigation of intraspecific variations of the chloroplast genome of Korean A. thaliana
. International Journal of Genomics 2020: 3236461.     Park, J. Xi, H. Kim, Y. Nam, S and Heo, K-I. 2020d. The complete mitochondrial genome of new species candidate of Rosa rugosa (Rosaceae). Mitochondrial DNA Part B Resources 5: 3435-3437.    Park, J. Xi, H. Park, J and Seo, BY. 2021d. A new mitochondrial genome of Sogatella furcifera (Horváth) (Hemiptera: Delphacidae) and mitogenome-wide investigation on polymorphisms. Insects 12: 1066.    Ronquist, F. Teslenko, M. van der Mark, P. Ayres, DL. Darling, A. Höhna, S. Larget, B. Liu, L. Suchard, MA and Huelsenbeck, JP. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539-542.    Shukla, N. Kuntal, H. Shanker, A and Sharma, SN. 2018. Mining and analysis of simple sequence repeats in the chloroplast genomes of genus Vigna
. Biotechnology Research and Innovation 2: 9-18.  Suh, H-J. Min, J. Park, J and Oh, S-H. 2021. The complete chloroplast genome of Aruncus dioicus var. kamtschaticus (Rosaceae). Mitochondrial DNA Part B Resources 6: 1256-1258.    Tatusova, TA and Madden, TL. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiology Letters 174: 247-250.   Wen, J and Jansen, RK. 1995. Morphological and molecular comparisons of Campsis grandiflora and C. radicans (Bignoniaceae), an eastern Asian and eastern North American vicariad species pair. Plant Systematics and Evolution 196: 173-183.   Wu, JY. He, SA and Gu, Y. 1990. Study on the consanguinity of Campsia grandiflora and C radicans
. Bulletin of the Nanjing Botanical Garden 1990. 46-50.
Wu, X. Li, H and Chen, S. 2021. Characterization of the chloroplast genome and its inference on the phylogenetic position of Incarvillea sinensis Lam. (Bignoniaceae). Mitochondrial DNA Part B Resources 6: 263-264.    Xiang, S. Liu, X-D. Sun, W-H. Lan, S-R. Liu, Z-J and Zou, S-Q. 2019. The complete chloroplast genome sequence of Euscaphis japonica (Staphyleaceae). Mitochondrial DNA Part B Resources 4: 3484-3485.    Xu, Z. Xin, T. Bartels, D. Li, Y. Gu, W. Yao, H. Liu, S. Yu, H. Pu, X. Zhou, J. Xu, J. Xi, C. Lei, H. Song, J and Chen, S. 2018. Genome analysis of the ancient tracheophyte Selaginella tamariscina reveals evolutionary features relevant to the acquisition of desiccation tolerance. Molecular Plant 11: 983-994.   Yang, J. Takayama, K. Youn, J-S. Pak, J-H and Kim, S-C. 2020. Plastome characterization and phylogenomics of East Asian beeches with a special emphasis on Fagus multinervis on Ulleung Island, Korea. Genes 11: 1338.    Yi, D-K and Kim, K-J. 2016. Two complete chloroplast genome sequences of genus Paulownia (Paulowniaceae): Paulownia coreana and P. tomentosa
. Mitochondrial DNA Part B Resources 1: 627-629.     Yoo, S-C. Oh, S-H and Park, J. 2021. Phylogenetic position of Daphne genkwa (Thymelaeaceae) inferred from complete chloroplast data. Korean Journal of Plant Taxonomy 51: 171-175.   Yu, H-C. Wu, J. Zhang, H-X. Zhang, H-S. Qiao, T-T. Zhang, J-X. Zhang, G-L. Sui, J. Li, L-W. Zhang, L-R and Lv, L-X. 2015. Antidepressant-like and anti-oxidative efficacy of Campsis grandiflora flower. Journal of Pharmacy and Pharmacology 67: 1705-1715.    Yuan, Y. Jin, X. Liu, J. Zhao, X. Zhou, J. Wang, X. Wang, D. Lai, C. Xu, W. Huang, J. Zha, L. Lui, D. Ma, X. Wang, L. Zhou, M. Jiang, Z. Meng, H. Peng, H. Liang, Y. Li, R. Jiang, C. Zhao, Y. Nan, T. Jin, Y. Zhan, Z. Yang, J. Jiang, W and Huang, L. 2018. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nature Communications 9: 1615.     Zerbino, DR and Birney, E. 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome research 18: 821-829.    Zhang, Z. Jiang, X. Chen, Y. Zhu, P. Li, L. Zeng, Y and Tang, T. 2019. Characterization of the complete chloroplast genome sequence of Conyza canadensis and its phylogenetic implications. Mitochondrial DNA Part B Resources 4: 2028-2030.  
|
|















