돌나물(돌나물과)집단의 유전적 변이: 남부지방 집단의 기원에 대한 암시
Notes on genetic variation in Sedum sarmentosum (Crassulaceae): Implications for the origin of southern Korean populations
Article information
Abstract
다년생 다육식물인 돌나물은 한반도 중남부에 흔하게 자란다. 이 종은 중국 고유종이며, 아마도 식용과 약 용 때문에 한반도에 도입되었을 것이라 추정된다. 만약 식물이 단일 공급원에서 한반도로 도입되었다면, 저자들은 남한집단에서 유전적 변이가 없거나 낮을 것으로 기대할 수 있다. 이 한두 곳에서 도입되었다면 알로자임 변이가 거의 없을 것으로 예상된다. 다른 한편으로 만약 식물종이 여러 곳에서부터 도입되었다면, 저자들은 이런 종이 높 은 수준의 유전적다양도를 유지하고 있을 것이라고 예측된다. 어떤 가설이 더 타당한지를 검증하기 위해, 저자들은 한반도 남부지방 10곳 집단을 대상으로 알로자임 변이를 조사하였다. 저자들은 조사된 15개의 모든 알로자임 유전 좌위에서 변이를 발견할 수 없었다. 그러나 두 동속종(기린초및 바위채송화)과 두 관련 종(둥근잎꿩의비름과 세잎 꿩의비름)은 중간 정도 및 높은 수준의 유전적 다양성을 유지하고 있음을 알 수 있었다(He 값이 각각 0.203, 0.144, 0.201 및 0.204). 저자들은 (한국남부지방, 자생지 및 다른 귀화된 돌나물이 불염이기에) 남부지역에 생육하는 돌나 물은 소수의 클론 분주체가 한번 도입된 후 무성적 번식에 의해 넓게 귀화되었을 것이라고 제안한다.
Trans Abstract
The succulent herbaceous perennial Sedum sarmentosum commonly grows in the southern part of the Korean Peninsula. It is a species native to China, most likely introduced into Korea due to its edible and medicinal uses. If plants were introduced from a single source, we would expect no or low levels of genetic variation in Korean populations. Alternatively, if plants were introduced from multiple sources, we would expect, in contrast, high levels of genetic diversity. To test which is more likely, we surveyed the degree of allozyme variation in ten populations of this species from southern Korea. We found that S. sarmentosum was monomorphic at all fifteen allozyme loci. However, two congeners (S. polytrichoides and S. kamtschaticum) and two related species (Hylotelephium ussuriense and H. verticillatum) maintain moderate to high levels of genetic diversity (He = 0.144, 0.203, 0.201, and 0.204, respectively). We suggest that southern Korean populations of S. sarmentosum likely descended from a single introduction of a few plants and then became naturalized exclusively via vegetative spreading (as plants in Korea, but also as occurs in other parts of its native and naturalized range, are sterile).
Sedum sarmentosum Bunge (Crassulaceae), commonly called “stringy stonecrop”, is a perennial succulent herb, 10–25 cm tall, that produces slender, creeping sterile shoots and erect flowering stems. Leaves are arranged in whorls of three, and inflorescences are cymes that produce pentamerous bright yellow flowers and ovoid seeds (ca. 0.5 mm) (Spongberg, 1978; Fu and Ohba, 2001; ‘t Hart and Bleij, 2003). Native to continental China, it occurs widely on rocky areas below 1,600 m mostly in central, eastern and north-eastern provinces (Anhui, Fujian, Gansu, Guizhou, Hebei, Henan, Hubei, Hunan, Jiangsu, Jiangxi, Jilin, Liaoning, Shaanxi, Shandong, Shanxi, Sichuan, and Zhejiang) (Fu and Ohba, 2001). Sedum sarmentosum has been used as medicinal and sometimes as a vegetable. By having an ornamental potential, it has been widely cultivated and naturalized in other parts of Asia (Japan, Korea, Taiwan, and north Thailand), but also in North America and many European countries (Ohwi, 1965, 2009; Sîrbu et al., 2011; Su and Lu, 2014). Largely owing to its vigorous vegetative spread and a high environmental tolerance, it has become an invasive plant even in some areas including Japan (Miyawaki and Washitani, 2004) as well as in some parts of the United States (e.g., West Virginia) (WVDNR, 2009).
Notably, Sedum sarmentosum has been recently reported as introduced in Taiwan (Su and Lu, 2014). Not documented in Flora of Taiwan (Tang and Huang, 1993), it was discovered along a roadside near Mt. Tatun in 2007. Su and Lu (2014) observed yellow flowers of S. sarmentosum along the road but no fruits were produced during a six-year monitoring, suggesting an asexual mode of reproduction. In South Korea, the first and the corresponding authors have observed flowers but failed to observe fruits in plants grown at a garden outside a greenhouse at Gyeongsang National University. As seems to occur in Taiwan, if plants were introduced from a single source and propagate vigorously by means of asexual reproduction, we would expect no genetic variation (assuming that the plant is monomorphic at genetic loci) in standing populations (e.g., Chung, 1999). Alternatively, if plant species were haphazardly or intentionally introduced from multiple source populations or areas, we would expect high levels of genotypic diversity (assuming that levels of genetic variation in source populations should be moderate or high) (e.g., Pappert et al., 2000).
More recently, neutral genetic markers (such as allozymes, microsatellites, amplified fragment length polymorphisms, etc.), have been employed to indirectly infer history of plant populations (Hamrick and Trapnell, 2011). Allozymes have been traditionally used in a variety of ways to better understand the origin and history of domestication or naturalization of plant species (Doebley, 1989; Pappert et al., 2000; Godt and Hamrick, 2001; Chung et al., 2014a). We have been interested in the origin of S. sarmentosum populations of southern Korea and used allozymes to test whether a single introduction or, alternative, multiple ones, is more likely.
Materials and Methods
We studied 10 populations, collecting a total of 329 samples (ranging from 12 in “E” to 53 in “F”) of S. sarmentosum from southern Korea (Fig. 1). Two sampling strategies were used; first, we sampled five populations from Juwangsan National Park (JNP) and its vicinity to assess levels of genetic variation at a small spatial scale (populations A to E). Outside JNP and its vicinity, we further collected samples from five locations (populations F to J) to study the genetic diversity at a large scale. The name of locations and sample sizes are included in the legend of Fig. 1. As S. sarmentosum exhibits extensive clonal growth, samples were collected at intervals of >2 m within each population to avoid biasing samples toward certain clonal ramets.
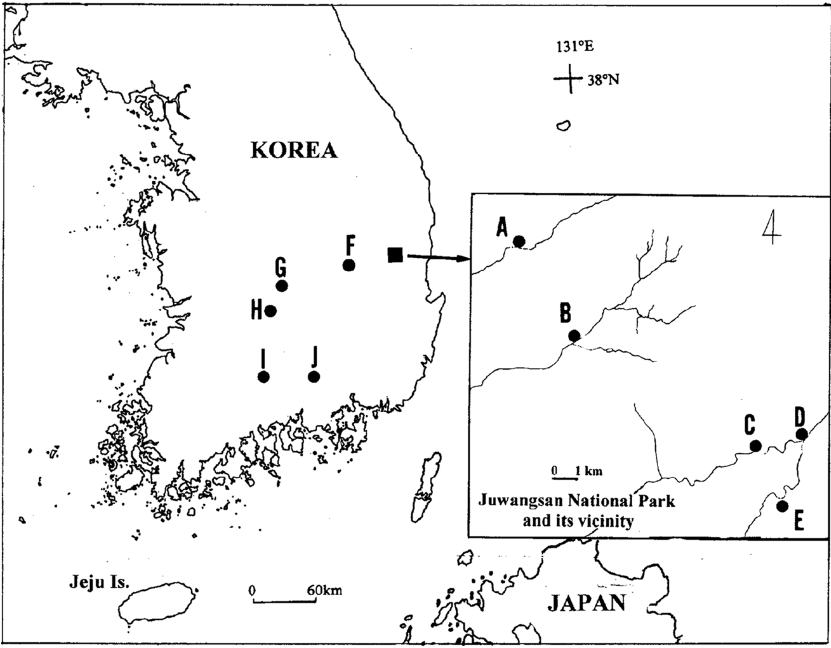
The location of the 10 populations of Sedum sarmentosum in Korea examined in this study: A–E in Juwangsan National Park (A, n = 18 and B, n = 24) and its vicinity (C, n = 29, D, n = 31, E, n = 12); F, Uisung gun, n = 53; G, Yeongdong gun, n = 48; H, Muju gun, n = 39; I, near Yeongcheon temple, Mt. Jiri, n = 52; J, Jinju city, n =23.
Using a phosphate-polyvinylpyrrolidone buffer (Mitton et al., 1979), we extracted enzymes and conducted horizontal starch-gel electrophoresis using 13% gels. We resolved 15 putative loci from ten enzyme systems using two buffer systems (Table 1). Electrophoretic methods and staining recipes are detailed in Chung et al. (2014b, 2016). We designated putative loci sequentially, with the most anodally migrating isozyme designated as 1, the next 2, and so on. We also designated different alleles per locus sequentially by alphabetical order (a, b, and so on).
We considered a locus to be polymorphic when two or more alleles were observed, regardless of their frequencies. We estimated the genetic diversity parameters within populations using the programs POPGENE (Yeh et al., 1999) and FSTAT (Goudet, 1995): percentage of polymorphic loci (%P), mean number of alleles per locus (A), observed heterozygosity (Ho), and Hardy-Weinberg (H-W) expected heterozygosity or Nei’s (1978) gene diversity (He).
Results and Discussion
All 15 allozyme loci surveyed were monomorphic in all populations (percentage of polymorphic loci %P = 0; total genetic diversity HT = 0) (Table 2). However, moderate to high levels of genetic diversity were detected in the congeneric S. kamtschaticum (%P = 66.7, A = 2.08, He = 0.203) (Chung et al., 2014b) (see Table 2) and S. polytrichoides (%P = 75, A = 2.25, He = 0.144) (Chung et al., 2016), and the nearly congeneric Hylotelephium ussuriense (%P = 83.3, A = 2.42, He = 0.201) (Chung et al., 2014b) and H. verticillatum (%P = 83.3, A = 2.00, He = 0.204) (Chung et al., unpubl. data). It should be noted that in these four species the loci surveyed were almost the same as in the present study (12 out of 15 loci), and the study area (JNP and its vicinity) is included within the one of S. sarmentosum (Fig. 1). In addition, despite buffer systems and the number of loci resolved are quite different from our study, five species of Sedum (S. aizoon, S. kamtschaticum, S. latiovalifolium, S. takesimense, and S. zokuriense) in South Korea also maintain high levels of withinpopulations genetic diversity [for more information, see Yoo and Park (2016) in this issue]. These comparisons clearly indicate that the genetic monomorphism of the studied Korean populations of S. sarmentosum is not due to the type of markers used, as allozymes are usually regarded as showing lower polymorphism compared to other markers such as microsatellites (Weising et al., 2005).

Summary of genetic diversity measures observed in Sedum sarmentosum, in two congeners, and in two species of the closely related genus Hylotelephium.
Our allozyme data, therefore, support the first prediction: Korean populations of S. sarmentosum have probably descended from a single introduction of few plants. Such patterns are not rare among invasive plants; for example, about 80% of the introduced populations of Eichhornia crassipes (Pontederiaceae) from all the world are composed of a single clone, suggesting a very few number of introduction events (Zhang et al., 2010). Similarly, many individuals of Alternanthera philoxeroides (Amaranthaceae) in their invasive range in China share the same multi-locus genotype even when the sampled populations are geographically very distant (Geng et al., 2016). However, multiple introductions cannot be excluded even in cases of low levels of genetic diversity, as the source populations can be genetically very similar (Lambertini et al., 2010), or the founding populations could have suffered from strong genetic bottlenecks or other processes leading to the loss of a large part of the genetic variation of the native populations (Rollins et al., 2013; Hagenblad et al., 2015). For the case of S. sarmentosum in Korea, it is unlikely that multiple introductions may have led to such monomorphism, as one would expect at least some genetic variation for a species that has a very large native area (a large part of China). Although no overall genetic assessments for the species in China are available, Xu et al. (2015) studied a population near Wuhan (Hubei Province) and found relatively high levels of microsatellite diversity. In addition, even though strong genetic bottlenecks at foundation would have produced a loss of genetic diversity, fixation for the same allele—as would have occurred for S. sarmentosum populations—is equally unlikely.
The apparent reliance on vegetative multiplication of this plant in Taiwan and in Korea, according to the observations of Su and Lu (2014) and our own observations, points towards a putative sterility for at least some of the forms of this species. According to a cytological study (Uhl and Moran, 1972), S. sarmentosum from the island of Honshu in Japan displayed considerable irregularity at meiosis and had about 72 chromosomes (n = ca. 36). This count might correspond to a tetraploid, given that the close congener S. angustifolium (both taxa are treated as the same species in Flora of China) (Fu and Ohba, 2001) is also 2n = 72, with a basic number that is considered to be x = 18 (Hsu et al., 1994). In addition, another close congener, Sedum lineare (‘t Hart and Bleij, 2003; Mayuzumi and Ohba, 2004), is also 2n = 72 (Uhl and Moran, 1972), suggesting that the basic number is certainly x = 18 for S. sarmentosum and its closely related species. A different chromosome number (2n = 58) has been, however, counted for samples from Shanghai (China) which was interpreted as an aneuploid (Hsu et al., 1994), that could likely be a neartriploid aneuploid, 2n = 3x + 4 = 58. We believe that the reason why there are no fruits in Taiwan and southern Korea, even with the formation of numerous flowers, is probably sterility (which is often caused by irregular meiosis or aneuploidy) (Khush, 1973; Ramsey and Schemske, 1998). Although we do not have any information about the chromosome number of S. sarmentosum in southern Korea, the allozyme monomorphism across the 10 studied populations suggests a probable sterility in the Korean plants. This could also be the situation of the species in North America, whose propagation in gardens was reported to be by means of stolons and not seeds, as the latter were not viable (Hsu et al., 1994). In China, its native area, Hsu et al. (1983) reported that some plants of S. sarmentosum set a few seeds (but others did not). The occurrence of putative fertile plants in China is also supported by the relatively large genetic variability for the assessed population of Hubei, which, in addition, harbors higher genetic variation than the studied individuals of the fertile S. lineare (Xu et al., 2015). We assume that S. lineare is fertile because there are many websites that sell seeds.
The discovery of S. sarmentosum in Taiwan in 2007 suggests that vegetative parts (stolons) would be carried by a traveler from mainland China to Taiwan, as the plants introduced to Taiwan did not set any fruit. Despite being common in Fujian (the province of mainland China that is geographically located in front of Taiwan), it is not surprising to have failed to find this species in Taiwan until recent times; after the establishment of the People’s Republic of China (in 1949), the cross-Strait travels were not allowed until 1987, when the ban was lifted (Guo et al., 2006). In the Korean Peninsula, in contrast, introduction of S. sarmentosum is probably much older due to two reasons: first, the species is not included in the list of the alien species that were introduced into the country between 1876 (the year that Korea was forced to open its doors as consequence of the Treaty of Ganghwa Island) and 2010 (Lee et al., 2011); and second, that the plant, commonly known as the dolnamul, has a long history as edible plant in Korea. Being used to make kimchi in the early spring, it was already listed in the Sallim Gyeongje (山林經濟), a book about farming that was written at the turn of the 17th and 18th centuries (KNA, 2014). A similar situation to Taiwan, however, would have occurred on Jeju Island (an island off the southern Korean coast) (Fig. 1); even though it commonly grows there at present, S. sarmentosum was hardly found on Jeju Island until mid-1970s (Kwack, 1976). Frequent travels between Jeju and continental Korea were not possible until the end of the darkest chapters of recent island’s history (the Japanese colonization of the island, that ended in 1945, and the 1947–1954 uprising) (Park, 2010), and, in fact, mass tourism did not start until the 1960s (Woo et al., 2013). Putting all these data together, it seems evident that S. sarmentosum was introduced from China into Korea and Taiwan (and perhaps Japan) through carrying parts of plants (not through seeds) by single (or very few) introduction events.
Our scenario of S. sarmentosum introduction in southern Korea is probably very similar to that found in Lycoris radiata var. radiata (Amaryllidaceae) from southern Korea (Chung, 1999), a plant that is also cultivated as ornamental and used in folk medicine. This variety is widely distributed in China, Korea, and Japan (Chung, 1999), and it spreads out vigorously by the formation of new bulbs, as it is a triploid (2n = 33, sterile, thus no seed set). The author, using 24 allozyme loci, examined eight populations from southern Korea and found no variation at those loci. Based on this finding, the author suggested that one or a few bulbs were introduced from China (or, secondarily, from Japan) to a temple in southern Korea. Later, the variety was naturalized in Korea via vigorous vegetative propagation by newly formed bulbs.
Sedum sarmentosum has been recognized as an important medicinal herb. It is often used in folk medicine (e.g., for the treatment of chronic hepatitis in China). More recently, it has been found to exhibit anti-angiogenic, anti-fibrotic, antiinflammatory, anti-nociceptive, anti-oxidant, anti-tumoral, and anti-viral properties (Kang et al., 2000; Jung et al., 2008; Mo et al., 2011; Bai et al., 2014). In China, high demand of S. sarmentosum for medicinal uses has led to overexploitation of the wild populations, driving them close to local extinction in the wild (Xu et al., 2015). It is also a plant increasingly used in roof greening in China and in Korea (Xiao et al., 2014; Lee et al., 2015), and, as noted above, a valued vegetable in the Korean cuisine. Although S. sarmentosum is commonly observed along roadsides and rocky areas in South Korea, it might be necessary to extensively survey genetic variation across the country, including several isolated islands by using highly variable markers to design guidelines for the preservation of its genetic resources (e.g., microsatellites) (Xu et al., 2015). As future studies, we suggest to further investigate the levels of genetic diversity across all the geographic range of this species in East Asia (China, Korea, and Japan), taking special care that all cytotypes are surveyed. The current chromosome counts (a putative tetraploid from Japan and an aneuploid from China) strongly suggest the existence of both diploid and triploid forms within the current range of S. sarmentosum, as triploids are generally produced by mating of diploids and tetraploids, and aneuploids may be either the result of errors at meiosis or from backcrossing of triploids to the diploid or tetraploid parents (Murray and Young, 2001; Henry et al., 2005).
Acknowledgements
This research was supported by the National Institute of Biological Resources, Republic of Korea to M.G. C. and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2013R1A1A2063524 and NRF-2013R1A1A 3010892 to M.G.C. and M.Y.C., respectively). This research was also supported by the grant no. 201630I024 (“Proyecto Intramural Especial, PIE” from the CSIC, Spain) to J.L.-P.
