The complete chloroplast genome sequence of Korean Neolitsea sericea (Lauraceae)
Article information
Abstract
The complete chloroplast (cp) genome sequence of Neolitsea sericea was determined by Illumina sequencing. The complete cp genome was 152,446bp in length, containing a large single-copy region of 93,796 bp and a small single-copy region of 18,506bp, which were separated by a pair of 20,072bp inverted repeats. A total of 112 unique genes were annotated, including 78 protein-coding genes (PCGs), 30 transfer RNAs, and four ribosomal RNAs. Among the PCGs, 18 genes contained one or two introns. A very low level of sequence variation between two cp genomes of N. sericea was found with seven insertions or deletions and only one single nucleotide polymorphism. An analysis using the maximum likelihood method showed that N. sericea was closely related to Actinodaphne trichocarpa.
The genus Neolitsea (Benth. & Hook.f.) Merr. (Lauraceae) consists of approximately 85 species of dioecious trees or shrubs and is distributed throughout tropical and subtropical Asia and Australia (Li et al., 2007; Qing et al., 2014). Among these species, Neolitsea sericea (Blume) Koidz., discussed in this study, was initially described as Laurus sericea (Blume, 1826) but has since been treated as a species belonging to the genus Neolitsea (Koidzumi, 1926). This species is an evergreen broadleaf tree and dioecious. The leaves are verticillate and elliptical with three pairs of lateral veins, and the flowers are yellowish white forming umbellate clusters without peduncle at the leaf axils. The fruit is a drupe and ripens to red in October of the following year. This species is very similar to N. aurata (Hayata) Koidz. and N. lunglingensis H. W. Li, but distinguished from these species in that the leaf blade is shortly acuminate and the fruit is globose (Huang and Werff, 2008; Kim, 2019).
Neolitsea sericea is distributed in China, Korea and Japan and is commonly found in the evergreen forests of Korea and Japan. The Korean Peninsula and the main island of Japan are also recognized as important regions because they represent the northern-most limit of its range (Ohashi et al., 2006; Lee and Choi, 2010; Lee et al., 2013). Due to these distributional characteristics, this species is recognized as a major indicator of climate change on the Korean Peninsula (Yun et al., 2014). In addition, the species is an important plant resource used as ornamental trees, timber, and in the creation of natural dyes and essential oils as treatments for inflammatory diseases (Jo et al., 2007; Yoon et al., 2010).
The first chloroplast (cp) genome sequence of this species was reported by Song et al. (2017) and was isolated in Zhejiang Province in China. In this study, we report the second complete chloroplast genome sequence of N. sericea from a sample collected on Jejudo Island in Korea. We also carry out comparative analyses between two plastomes to recognize the infraspecific variation.
Materials and Methods
Plant material
The plant materials were sampled from Bonggae-dong (33°25′37.8″N, 126°38′39.2″E), Jeju-si, Jeju-do, and a voucher specimen was deposited in the Sangji University Herbarium (SJUH; Kyeong-Sik Cheon, cheonks@sangji.ac.kr), with voucher number SJUH000861.
DNA extraction and cp genome determination
Total DNA was extracted from fresh leaf using a DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA). The Paired-end library was constructed with a TruSeq DNA Sample Preparation Kit (Illumina Inc., San Diego, CA, USA). Genomic DNA sequencing was performed using the Illumina MiSeq (Illumina Inc.) platform to produce 3,438,412 raw reads with a length of 301 bp.
The assembly and annotation of the cp genome were accomplished using the Geneious Prime v2020.1.3 (Biomatters Ltd, Auckland, New Zealand). For correct gene annotation, we also compared each gene to four published complete chloroplast genomes of Lauraceae: Actinodaphne trichocarpa C. K. Allen (GenBank accession no. MF939342), Lindera pulcherrima (Nees) Hook.f. (GenBank accession no. MN453268), L. chunii Merr. (GenBank accession no. NC045254), and L. aggregata (Sims) Kosterm. (GenBank accession no. NC045252). The tRNAs were confirmed using tRNAscan-SE v2.0 (Chan and Lowe, 2019). A circle cp genome map was drawn using the OGDRAW program (Greiner et al. 2019).
Phylogenetic analysis
To construct the phylogenetic tree, a total of 74 protein-coding genes (PCGs) of 50 species were aligned using the MAFFT (v7.450) program (Katoh and Standley, 2013). Forty-eight Lauraceae accessions were selected as the ingroups, and two Calycanthaceae species, Chimonanthus nitens Oliv. (MH377057) and C. praecox (L.) Link (MH377058), were chosen as outgroups. We selected the GTR+I+Γ model as the best substitution model according to jModelTest version 2.1.10 (Darriba et al., 2012). Maximum likelihood (ML) analyses were performed using PAUP* 4.0a168 (Swofford, 2002) with 1,000 bootstrap replicates.
Results and Discussion
A circular form of the complete chloroplast genome of N. sericea (MW465540) is 152,446bp in length with 39.2% G+C content, composed of a large single-copy (LSC) region of 93,796bp, a small single-copy (SSC) region of 18,506bp, and two inverted repeat (IR) regions of 20,072bp (Fig. 1). The whole genome of Korean N. sericea was 4 bp longer than that of the first cp genome (152,442 bp); the LSC region was 7 bp shorter, SSC region was 1 bp longer, and the IR regions were 5 bp longer. Also, the LSC region of the second cp genome was 7 bp shorter, and its SSC and IR regions were 1 bp and 5 bp longer than the corresponding regions of the Chinese cp genome (MF939341). The plastid genome contains a total of 112 unique genes consisting of 78 PCGs, 30 transfer RNAs, and four ribosomal RNAs. Additionally, twelve complete genes are duplicated in the IR regions. Among the 112 unique genes, eighteen (atpF, clpP, ndhA, ndhB, petB, petD, rps12, rpl16, rpoC1, rps12, rps16, ycf3, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one or two introns.
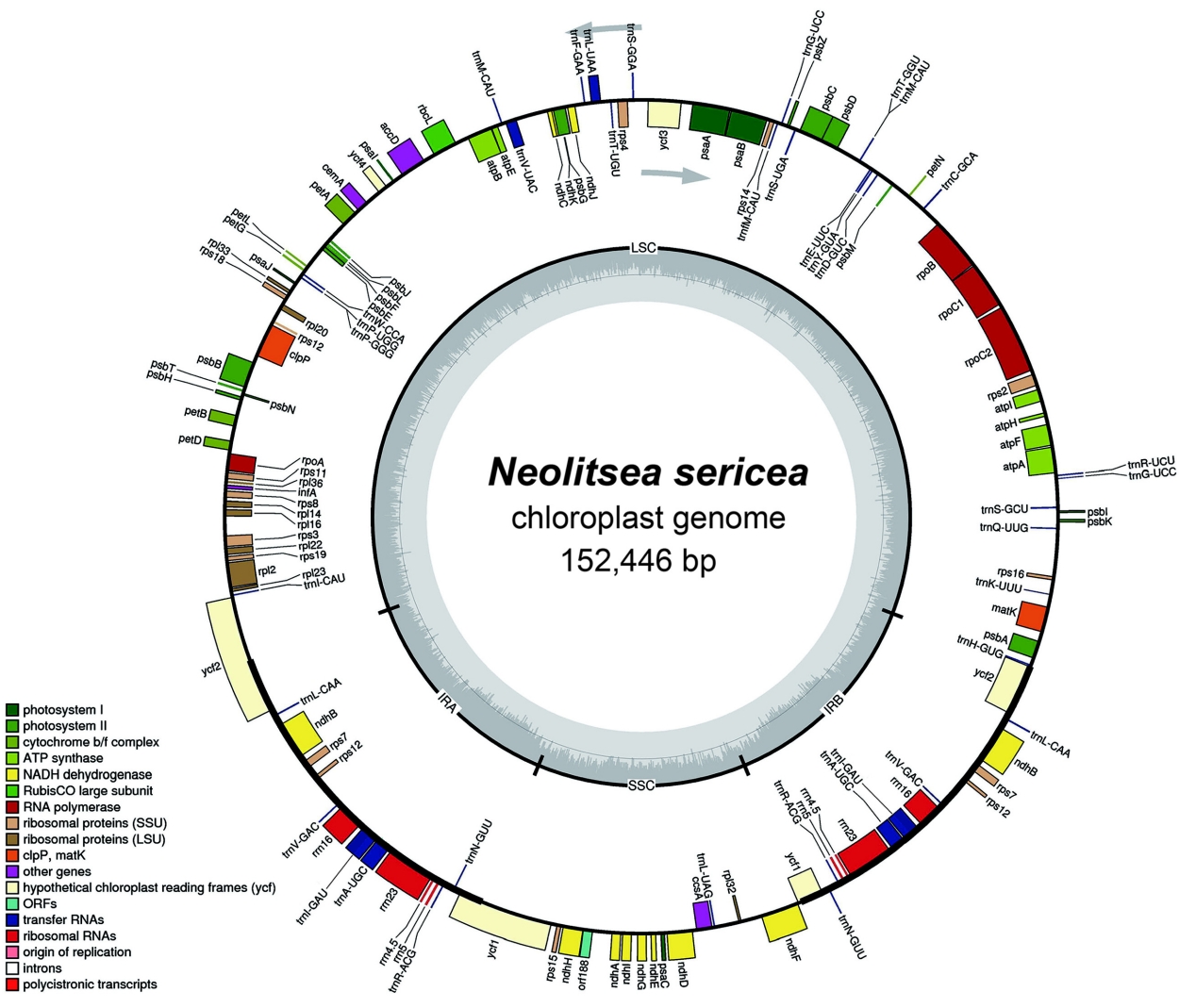
Structural map of the chloroplast genome in Neolitsea sericea. Genes inside the circle are transcribed clockwise, genes outside are transcribed counter-clockwise. The dark gray inner circle corresponds to the GC content, the light-gray to the AT content.
Based on the pairwise alignment of the two cp genomes, seven insertions or deletions (INDELs) and one single nucleotide polymorphism (SNP) were identified (Table 1). Among these, six indels and one SNP were in the LSC, and only one INDEL was correspondingly identified in the SSC. The infraspecific cp genome variation in this species is significantly lower than those in other land plants: 121 SNPs and 781 INDELs in Pyrus ussuriensis Maxim. (Cho et al., 2019), 84 SNPs and 125 INDELs in Pseudostellaria palibiniana Takeda (Kim et al., 2019), 69 SNPs and 660 INDELs in Marchantia polymorpha L. (Kwon et al., 2019), 78 SNPs and 643 INDELs in Camellia japonica L. (Park et al., 2019), 16 SNPs and 49 INDELs in Viburnum erosum Thunb. (Choi et al., 2020), 258 SNPs and 542 INDELs in Agrimonia Pilosa Ledeb. (Heo et al., 2020), 7 SNPs and 4 INDELs in Veronica nakaiana Ohwi (Lee et al., 2021), 33 SNPs and 662 INDELs in Campanula takesimana Nakai (Park et al., 2021), and 69 SNPs and 650 INDELs in Daphne genkwa Siebold & Zucc. (Yoo et al., 2021).
The ML tree (Fig. 2) formed five clades. The first clade consisted of the genera Beilschmiedia Nees, Cryptocarya R. Br. and Eusideroxylon Teijsm. & Binn. and formed the most basal part. The second clade is made up of Neocinnamomum H. Liu and Caryodaphnopsis Airy-Shaw. The third clade and fourth clade consist of four genera (Alseodaphne Nees, Persea Mill., Machilus Nees, and Phoebe Nees) and three genera (Nectandra Rol. Ex Rottb., Cinnamomum Schaeff., and Sassafras L. ex Nees), respectively. The last clade consists of the remaining genera excluding those mentioned above with Lindera (Adans.) Thunb. being paraphyletic. Also, N. sericea is closely related to A. trichocarpa with a high support value (bootstrap value = 100).
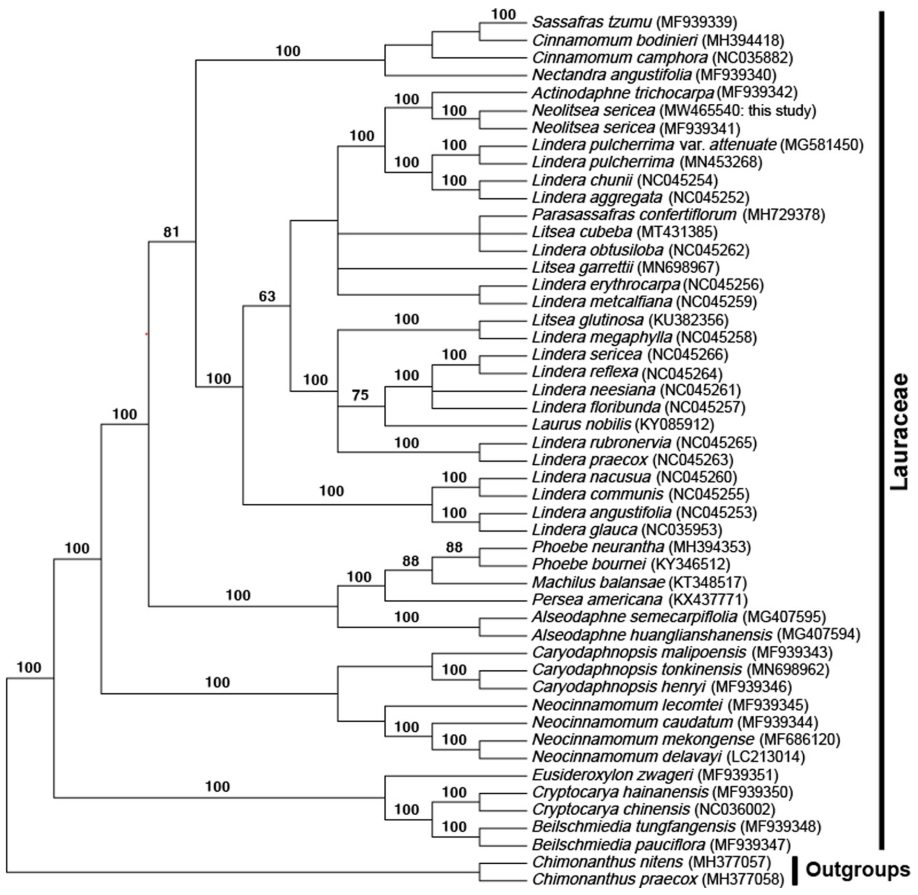
The maximum likelihood tree based on 74 protein-coding genes from 48 Lauraceae species and two outgroups. Numbers on branches indicate bootstrap value greater than 50%.
A comparative analysis of the cp genome sequences between the Korean and Chinese N. sericea in this study provides useful information to understanding of the evolution and diversification of this species. This study will provide important information for further studies of the ecology and genetics/genomics of this species.
Acknowledgements
This study was performed with the support of the Sangji University and Graduate School of Sangji University.
Notes
Conflict of Interest
The authors declare that there are no conflicts of interest.

