The complete chloroplast genome sequence of Hibiscus sabdariffa (Malvaceae)
Article information
Abstract
Hibiscus sabdariffa L., (roselle) in the Malvaceae family is an erect subshrub known to be native to India and Malaysia. It is widely used as a food or tea material around the world, and its therapeutic effects have been widely studied. In this study, the sequencing of the complete chloroplast genome of H. sabdariffa was carried out. The result indicates a genome size of 162,428 bp, which is composed of a large single copy of 90,327 bp, two inverted repeats of 26,242 bp each, and a small single copy of 19,617 bp. Overall, a total of 131 genes were predicted, including 86 coding sequences, 37 tRNAs, and 8 rRNAs. According to a phylogenic analysis, it was clearly distinguished from outgroups such as other species of the genus Hibiscus used in the analysis.
INTRODUCTION
Malvaceae consists of 244 genera and 4,225 species (Christenhusz and Byng, 2016). In the genus Hibiscus, more than 250 species are distributed from the temperate to tropical climate regions (Mandaji et al., 2022), and these species include herbs, shrubs, or trees (Abdullah et al., 2020). Plants of the genus Hibiscus have been studied and used not only for ornamental purposes, but also as functional foods and medicines (Abdelhafez et al., 2020).
Hibiscus sabdariffa L., commonly known as roselle, is an herbaceous subshrub and can reach up to 2.5 m in height, and generally possesses red stems and calyces (Morton, 1987; Da-Costa-Rocha et al., 2014; Bule et al., 2020). Roselle is known to be native to India and Malaysia, but it has been cultivated and spread in countries with tropical and subtropical climates regions (Izquierdo-Vega et al., 2020). Roselle has a variety of uses: the seeds are used as a source of dietary fibres, antioxidants, and edible oil. The dried calyx is used in beverages and teas known as carcade that are effective against chronic non-communicable diseases (Dhar et al., 2015; Montalvo-González et al., 2022). Roselle has been mainly studied on its utility in various industries, but few on genetic characteristics and structure (Sánchez-Mendoza et al., 2008; Montalvo-González et al., 2022).
Chloroplast is an organelle that performs primarily photosynthesis, and its genome sequence is well conserved, so it is a major material utilized in the studies of species classification, differentiation and evolutionary process (Liu et al., 2019; Cheng et al., 2020; Kim et al., 2021). In this study, the chloroplast complete genome sequence assembly of roselle was peformed for the first time to be used as an important material for further studies on the evolution and biodiversity of the species and among the species of Hibiscus.
MATERIALS AND METHODS
Fresh leaves of H. sabdariffa were sampled in National Institute of Forest Science, Suwon, Korea. Total DNA was extracted by GeneAll Genomic DNA Purification Kit (GeneAll Biotechnology, Seoul, Korea). DNA library was constructed using TruSeq Nano DNA Kit with a protocol according to the Sample Preparation Guide provided by the manufacturer (Macrogen Inc., Seoul, Korea). Genome sequencing was performed on the Illumina NovaSeq 6000 platform with 151 bp read size and paired-end type (Macrogen Inc.).
The chloroplast complete genome was assembled using NOVOPlasty v.4.3.1, an organelle assembler (Dierckxsens et al., 2017). To increase assembly reliability, repeated assemble work was performed with K-mer 21, 23, 25, 27, 29, 31, 33 based on two reference genome sequences, H. syriacus, NC_026909 (Kwon et al., 2016) and H. sinosyriacus, MZ_367751. Comparative sequence verification and error correction were carried out by manually. Gene annotation was performed using BLATN, BLATX, and Chloe v. 0.1.0 in GeSeq, an annotator of organelle genomes (Tillich et al., 2017). A circular map of the chloroplast complete genome was drawn by OGDRAW v. 1.3.1 (Greiner et al., 2019). Microsatellite analysis was carried out by MIcroSAtellite (MISA) v. 2.1 with a default setting (Beier et al., 2017).
For phylogenic analysis, total nine chloroplast genome sequences of Malvaceae were used, including H. sabdariffa and two outgroups, Gossypium raimondii and G. trilobum. Sequences were aligned using the Clustal Omega v. 1.2.4, alignment program (Sievers et al., 2011). The phylogenetic tree was reconstructed using the Maximum Likelihood method with the JTT matrix-based model and 1,000 bootstrap replicates in MEGA v. 11.0.11 (Tamura et al., 2021).
RESULTS AND DISCUSSION
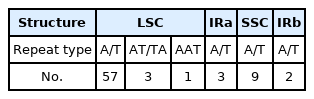
A total of 159,895,334 reads were produced in assembly work. Of these, 1,768,742 reads, about 9% of total reads mapped to the reference genomes, H. syriacus and H. sinosyriacus and the average organelle coverage of total reads compared to overlapped sequences was 9162x. Finally, the chloroplast complete genome of H. sabdariffa was assembled. Its total genome size was 162,428 bp. The chloroplast genome of the species was registered in NCBI’s GenBank with accession number, MZ_522720. The associated BioProject, BioSample, and SRA numbers are PRJNA_789603, SAMN_24146078, and SRR_17253144, respectively. The genome is composed of four regions: large single-copy (LSC), two inverted repeats (IRs), and small single-copy (SSC). LSC contains 90,327 bp, while IRa and IRb contain 26,242 bp each, and SSC has 19,617 bp (Fig. 1). In total, 131 genes comprising 86 coding sequences, 37 tRNAs, and 8 rRNAs are predicted. There were 75 simple sequence repeats (SSRs): 71 of them were monomeric repeats (Table 1). In particular, 81.3% of the total SSRs were found in LSC, and dimeric and trimeric repeats were also found in the LSC region. Nine monomeric repeats were found in the SSC region, and the five were in the IRs region.

A circular map and annotations of chloroplast complete genome of Hibiscus sabdariffa drawn by OGDRAW v. 1.3.1 and annotated by BLATN, BLATX, and Chloe v. 0.1.0 in GeSeq organelle annotation web tool.
Results of phylogenetic analysis showed that the genomes of Hibiscus formed a well-supported clade (Fig. 2). Among the seven Hibiscus species in the phylogenetic tree, H. sabdariffa is sister to all other species of Hibiscus. The complete chloroplast genomes of Hibiscus are too few to correctly infer the phylogenetic relationships of the members of the genus, considering the number of species of the genus. However, our phylogenetic analysis suggests that H. sabdariffa is distinct. The complete chloroplast genome of H. sabdariffa determined in this stduy will provide useful information for further studies on evolution and biodiversity with Hibiscus species and Malvalceae, which contain many economically important plants.
Acknowledgements
This work was supported by the National Institute of Forest Science (NiFoS) grant funded by the project number FG0403-2018-01.
Notes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.