The complete chloroplast genome sequence of Dracocephalum rupestre (Lamiaceae)
Article information
Abstract
Dracocephalum rupestre Hance is a perennial herb distributed across China, Mongolia, and Korea. This study reports the first complete chloroplast genome sequence of D. rupestre. The plastome is 151,230 bp long and exhibits a typical quadripartite structure comprising a large single-copy region of 82,536 bp, a small single-copy region of 17,408 bp, and a pair of identical inverted repeat regions of 25,643 bp each. It contains 130 genes, comprising 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis of D. rupestre and related species of Lamiaceae showed that the genus Dracocephalum is a monophyletic group, and D. rupestre is most closely related to D. psammophilum.
INTRODUCTION
The family Lamiaceae comprises more than 7,000 species distributed worldwide and is the sixth largest family of angiosperms (Li et al., 2016). The genus Dracocephalum L., also known as dragonheads, comprises approximately 70 species that are native to temperate regions in the Northern Hemisphere (Heydari et al., 2019). This genus can be readily distinguished from related genera of the subtribe Nepetinae based on the presence of thickened folds between the base of the calyx teeth and the general presence of aristately toothed bracteoles (Chen et al., 2021).
Dracocephalum rupestre Hance is a perennial herb distributed in the alpine meadows of China, Mongolia, and Korea that grows to a height of 30 cm and generally possesses purple-blue flowers (Lee, 1980; Zhu et al., 2018). D. rupestre is widely known as a traditional herbal medicine for the treatment of various diseases such as headache, fever, liver toxicity, and hepatitis (Zhu et al., 2018). Also, in China, the leaves of D. rupestre are used for manufacturing tea beverage, that called Maojian tea (Gao et al., 2022), and it is also used as an ornamental herb for large purple-blue flowers (Li and Hedge, 1994). In Korea, D. rupestre is a vulnerable species and limitedly distributed in the cracks of rocks in limestone areas in Gangwon-do, and less than 10 habitats are known; therefore, mine development, including the operation of quarries, may reduce the extent of its habitat (Korea National Arboretum, 2021).
The chloroplast (cp) is a semiautonomous organelle that performs primarily photosynthesis and plays an important role in biochemical processes in land plants (Wang et al., 2018; Kwon et al., 2022). The chloroplast genomes of most terrestrial plants have highly conserved circular structures consisting of large single-copy (LSC), small single-copy (SSC), and two large inverted repeats (IRs), range in size from 115 to 165 kb, and consist of 130 genes (Lee et al., 2007; Zhang et al., 2014; Song et al., 2017). The chloroplast genome has been used for phylogenetic analysis because of its highly conserved structure and slower evolution compared to nuclear genomes (Bi et al., 2018). Additionally, genetic information of the chloroplast genome is maternally inherited in most angiosperms (Song et al., 2017). Recently, many systematists have used whole plastid genomes for phylogenetic analysis among groups of plants whose evolutionary relationships remain unresolved (Gitzendanner et al., 2018).
Herein, we report the complete chloroplast genome sequence of D. rupestre. Additionally, we analyze its phylogenetic relationship with other Lamiaceae species using comparative analysis of completed chloroplast genomes to provide useful data for future studies of this taxon.
MATERIALS AND METHODS
Dracocephalum rupestre was collected from Singi-myeon, Samcheok-si, Gangwon-do, South Korea (37º19′23.9″N, 129º00′18.8″E). The collected specimen was prepared and deposited at the Herbarium of the Korea National Arboretum under voucher number ESK21-346.
Total genomic DNA was extracted using a DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, USA) and visualized on agarose gel (1%). Following library preparation, the prepared libraries were sequenced on the MiSeq platform (Macrogen, Seoul, Korea). A total of 7,100,628 paired-end reads were obtained and 219,741 reads were identified as chloroplast genome sequences. The average coverage depth of these chloroplast reads was 97.6×. Generated reads were assembled with a de novo strategy using Geneious Prime v2022.1.1 (Biomatters Ltd., Auckland, New Zealand) and annotated using the GeSeq tool (Tillich et al., 2017). A circular map of the complete genome of D. rupestre chloroplast was drawn using OrganellarGenomeDRAW v. 1.3.1 (OGDRAW) (Greiner et al., 2019).
To confirm the phylogenetic position of D. rupestre within Lamiaceae, whole chloroplast genome sequences of 24 species (six Dracocephalum species and 18 representative species belonging to Lamiaceae) and one outgroup taxon (Verbena officinalis; NC_056142) were downloaded from NCBI GenBank. For phylogenetic analysis, 79 protein-coding DNA sequences (CDSs) from 26 chloroplast genomes (including the outgroup) were selected and aligned using MAFFT in Geneious Prime v2022.1.1 (Biomatters Ltd.). The phylogenetic tree was constructed using the maximum likelihood (ML) method with 1,000 bootstrap replicates in IQ-TREE v.1.6.7 (Nguyen et al., 2015). The best-fit model was also determined as GTR + F + R3 (general time reversible model with unequal rates and unequal base frequency + empirical base frequencies + FreeRate model with 3 categories) through IQ-TREE v.1.6.7 (Nguyen et al., 2015).
RESULTS AND DISCUSSION
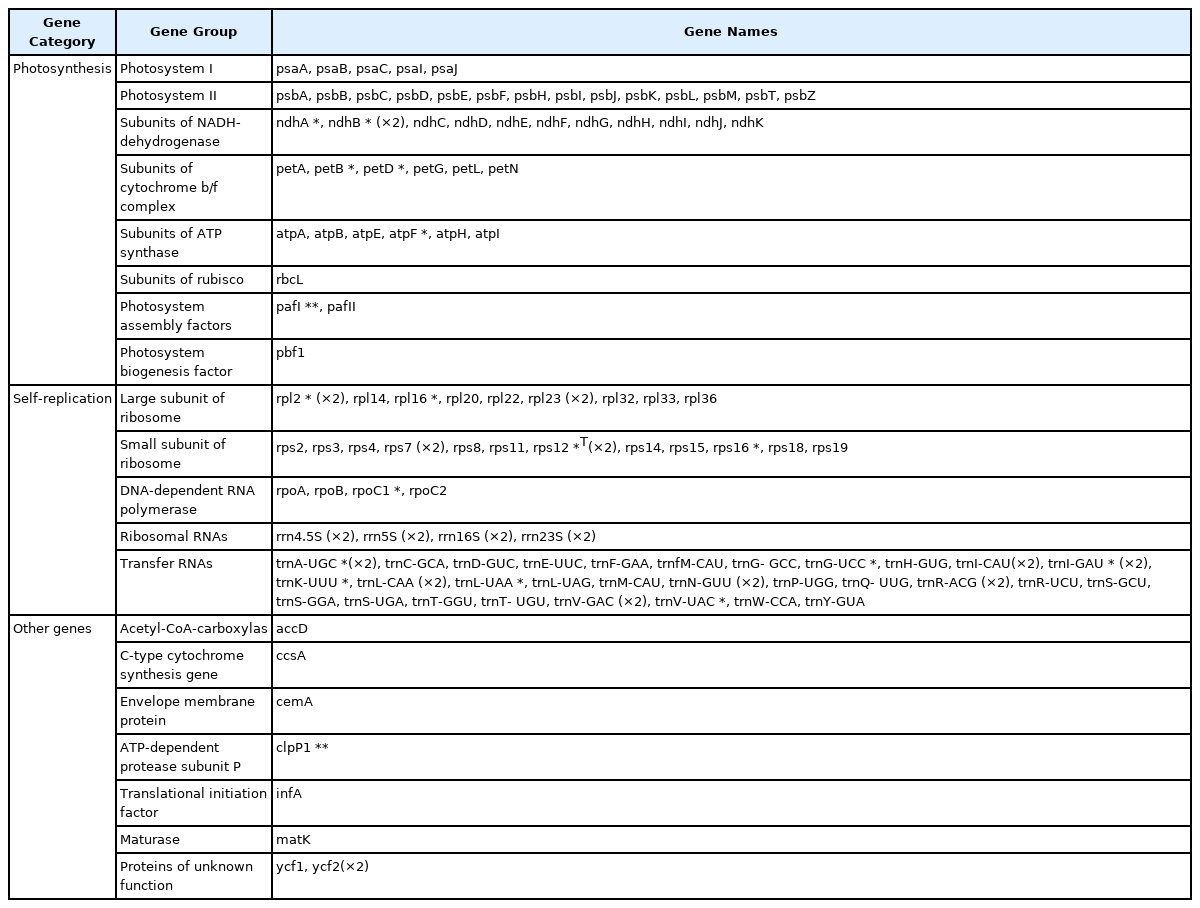
The complete chloroplast genome of D. rupestre (GenBank accession number: OP605968) was 151,230 bp in length with an overall GC content of 37.8%, consisting of an LSC region of 82,536 bp, an SSC region of 17,408 bp, and a pair of IR regions of 25,643 bp each (Fig. 1). It contains 130 genes, comprising 85 protein-coding genes, 37 tRNAs, and eight rRNAs. A total of 17 genes were duplicated in the IR region, including seven tRNA genes (trnA-UGC, trnI-CAU, trnIGAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), four rRNA genes (rrn4.5S, rrn5S, rrn16S, and rrn23S), and six protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12, and ycf2), of which the ycf1 gene at the SSC/IRb border is considered a pseudogene as it is only partially duplicated. Ten protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, and rps16) contained one intron, and two protein-coding genes (clpP1 and pafI) contained two introns (Table 1). rps12 is a trans-spliced gene with exon1 located in the LSC region and exon2 and exon3 located in IR regions.
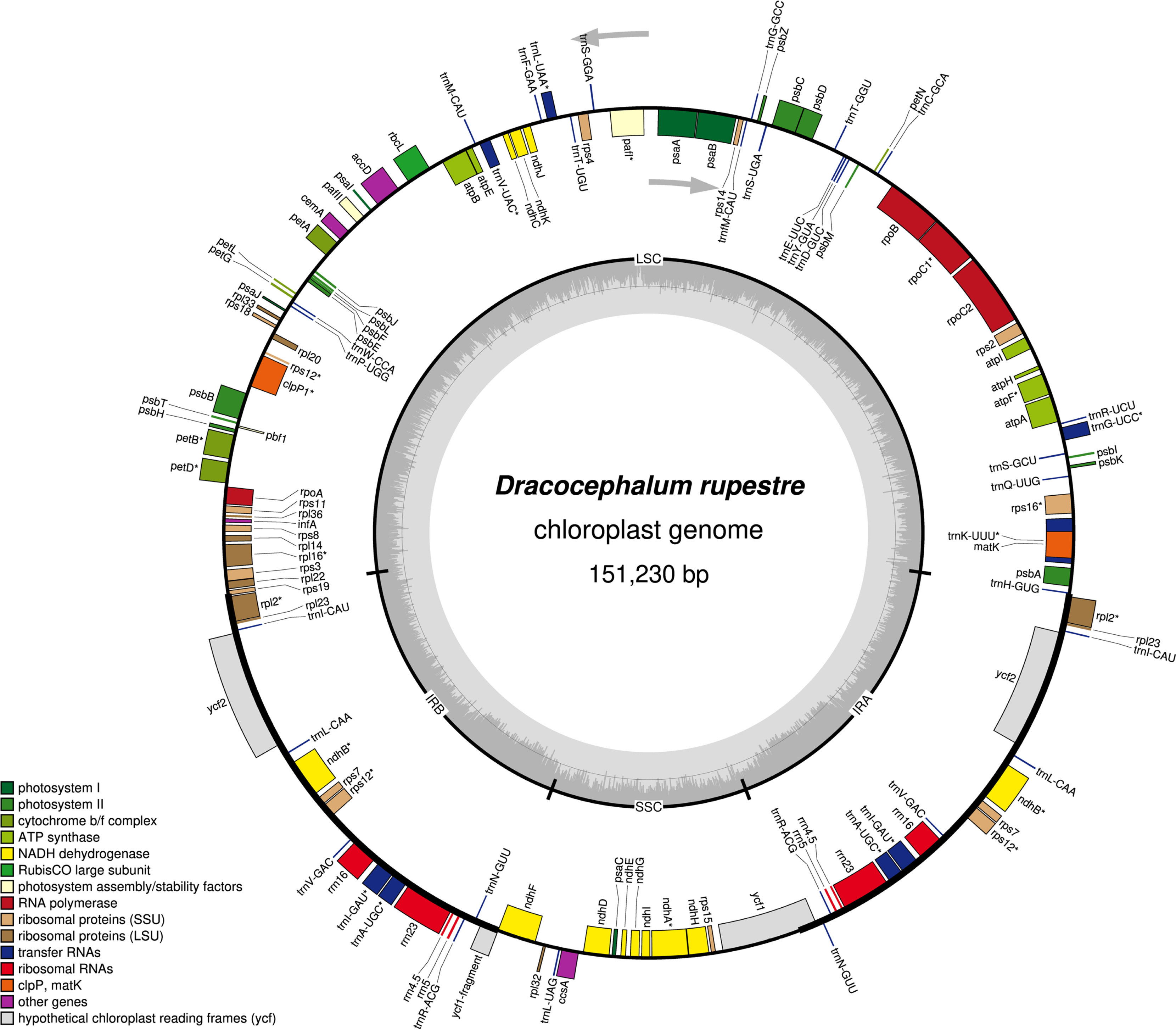
A circular map and annotations of complete chloroplast genome of Dracocephalum rupestre drawn by OGDRAW v. 1.3.1. Genes drawn inside and outside of the circle are transcribed in clockwise and counterclockwise, respectively. Large single-copy (LSC), small single-copy (SSC), and inverted repeat (IRA and IRB) are indicated.
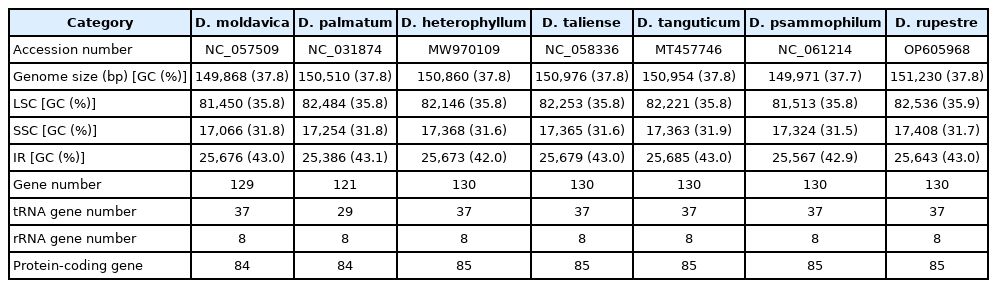
Compared to the previously published six species of the genus Dracocephalum, the range of chloroplast genomes of Dracocephalum species is from 149,868 bp (D. moldavica) to 150,976 bp (D. taliense). Regarding of gene order and contents, they have identical gene order. But D. heterophyllum, D. psammophilum, D. tanguticum, and D. taliense have identical gene contents with D. rupestre, D. moldavica have 129 genes because of loss of rps2 gene, and D. palmatum have less gene contents (121 genes) (Table 2).
Phylogenetic analysis using 79 CDSs revealed that Lamiaceae form a monophyletic group and can be divided into two clades. The first clade includes species of the subfamily Lamioideae, Scutellarioideae, and Ajugoideae. The second clade comprises species in the subfamily Nepetoideae. The topology of the phylogenetic tree is consistent with that of a previous phylogenetic study based on five combined cpDNA regions (Li et al., 2016). Additionally, the genus Dracocephalum is monophyletic (100% bootstrap support) and sister to the clade consisting of the closely related genera Mentha, Clinopodium, and Thymus (Fig. 2). All seven species of the Dracocephalum used in this study are included in subgenus Dracocephalum (Budantsev, 1987; Chen et al., 2022).
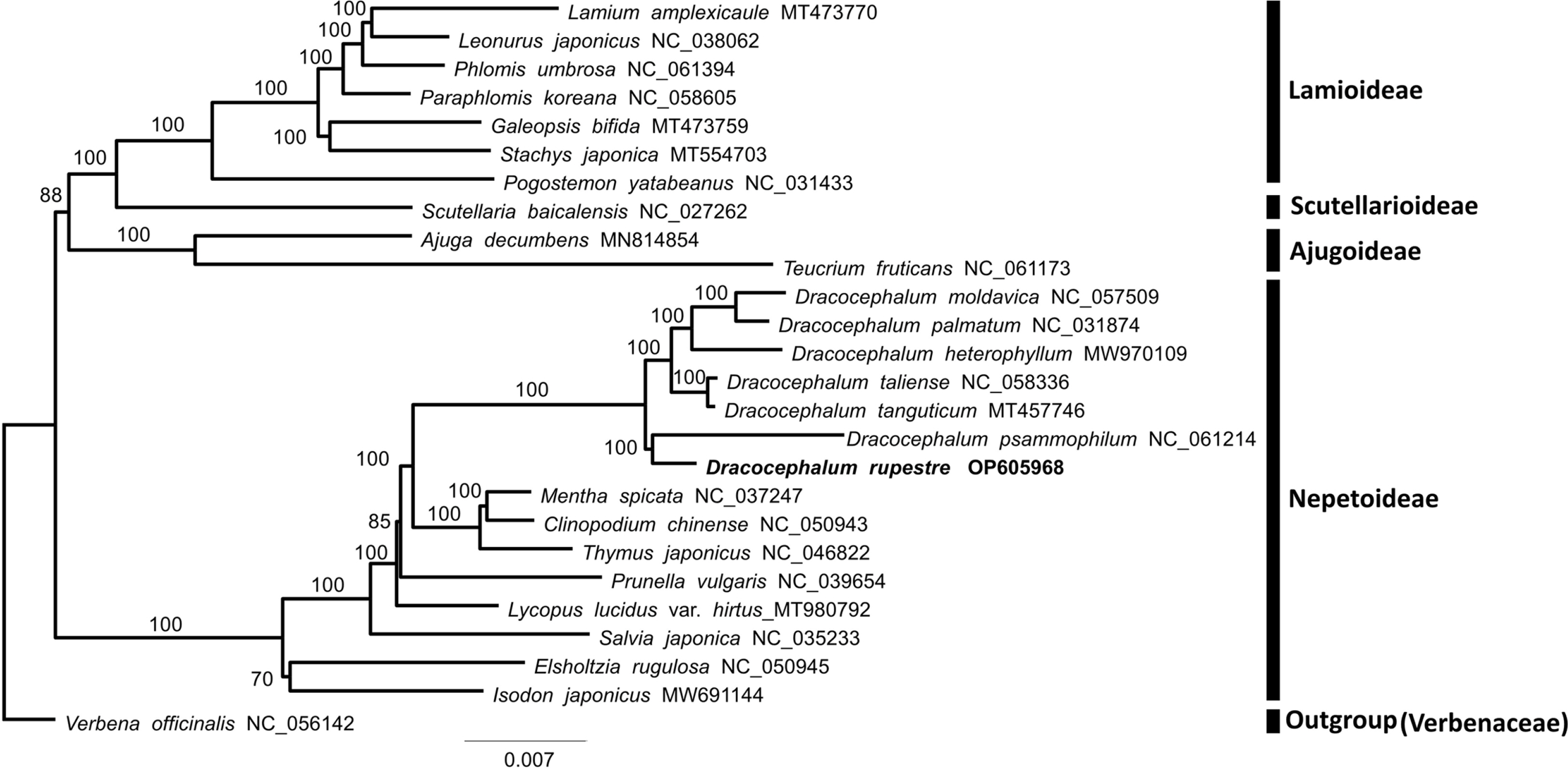
Maximum likelihood (ML) tree based on 79 protein-coding DNA sequences (CDSs) of Dracocephalum rupestre Hance, 24 species of Lamiaceae, and Verbena
Furthermore, the ML tree indicated that D. rupestre is closest to D. psammophilum. Compared to the chloroplast genome of D. rupestre and D. psammophilum, the pairwise identity percent is 96.5%. Although they have identical gene order and contents, the chloroplast genome of D. rupestre is 1,259 bp longer than that of D. psammophilum (149,971 bp, NC_061214). Dracocephalum psammophilum is a subshrub that is distributed in deserts of China that grows to a height of 7 cm and generally has purple-blue flowers. Dracocephalum rupestre can be morphologically distinguished from D. psammophilum by stems 30 cm long, basal leaves present, calyx 2–2.4 cm, and corolla 3.8–4 cm (Li and Hedge, 1994). But the most recent phylogeny study of the genus Dracocephalum based on two nrDNA regions (ITS and ETS), two low-copy nuclear markers, and four cpDNA regions show that D. rupestre and D. psammophilum are divided into different clades, and reveal relatively distant phylogenetic relationships (Chen et al., 2022). Despite their morphological and molecular differences, the close relationship between the two species in this study is due to the lack of the complete chloroplast genomes of Dracocephalum to reconstruct the phylogenetic relationships of the genus. Further studies are necessary to recognize fully resolved phylogenetic relationships within the genus.
The complete chloroplast genome of D. rupestre is reported for the first time in this study. The chloroplast genome sequence of D. rupestre determined herein will provide useful information for future studies to understand the phylogenetic and evolutionary relationships of Lamiaceae species.
Acknowledgements
This study was supported by a grant from the Korea National Arboretum (KNA1-1-13, 14–1)
Notes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.