Gnetum (Gnetaceae, Gnetales) is consisted of about 33 species of woody climbers and two tree species, distributed in tropical areas of West Africa, Central to South America, and Asia, with most species diversity concentrated in tropical and subtropical Asia (Markgraf, 1929, 1951, 1965; Kubitzki 1990; Steveonson and Zanoni, 1991; Price, 1996; Won and Renner, 2006; Biye et al. 2014). Won and Renner (2003, 2005a, 2005b, 2006) have studied the phylogeny, biogeography, and evolution of Gnetum, by analyzing sequences of mitochondrial nad1 gene intron, chloroplast (cp) rbcL and matK gene, trnL intron and adjacent spacers, and nuclear (nu) ribosomal ITS and Leafy gene. Hou et al. (2015) sequenced nuclear 18S and 26S ribosomal regions, together with cp matK gene, of 58 Gnetum accessions to test the classification schemes previously suggested, primarily based on the framework of Won and Renner (2006). However, proper identification of Gnetum species has been difficult, since Gnetum is dioecious gymnosperm with broad leaves, which renders scarcity of diagnostic characters without reproductive structures. Especially seeds are indispensable for identification. Because of this, there has been confusion and uncertainties regarding identity and biodiversity assessment of Gnetum.
DNA barcoding has been applied to identify biological specimens, by amplifying and sequencing short gene sequence (Hebert et al., 2003; Kress et al. 2005). DNA barcoding has been successfully applied not only to species identification, but also to ecological and community-level studies (Fazekas et al., 2008; Lahaye et al., 2008; Valentini et al., 2008; Kress et al., 2009; Fazekas et al., 2012; Taylor and Harris, 2012; Ajmal Ali et al., 2014; Kress et al., 2015). Several DNA regions of chloroplast and nuclear genome has been applied for DNA barcoding of land plants, with rbcL+matK has been recommended as a core barcode (CBOL Plant Working Group, 2009; Li et al., 2011), supplemented with trnH-psbA or nu ITS (China Plant BOL Group, 2011). One of the latest DNA barcode markers for land plants has been cp ycf1 (Dong et al., 2015). For Gnetum, most of the sequencing was done by Won and Renner (2003, 2005a, 2005b, 2006) and Hou et al. (2015). Although Won and Renner (2006) have developed primer combinations to amplify and sequence the whole cp trnK intron regions including matK gene, only 12 samples were sequenced by Won and Renner (2006) and Hou et al. (2015) added 37 matK sequences of Gnetum. The matK sequences of Gnetum provided more phylogenetic information, since 3 times more sequence divergences were observed for matK gene compared to rbcL gene (Won and Renner, 2006). Although the length of the full matK gene is 1554–1557 bp for Gnetum (Won and Renner, 2006), portion of the gene, which contains enough informative sites for species recognition, can be utilized as barcoding marker.
Since June 2010, we have been surveying and inventorying seeds plants of Cambodia, as a cooperative project between the National Institute of Biological Resources, Ministry of Environment, Republic of Korea and the Forest Administration, Government of Cambodia. During the field work, we have encountered and collected Gnetum species, as it is a common component of tropical evergreen forest. However, not all the materials are with seeds or reproductive structure, which are critical for identification, and even worse the herbarium record for Cambodian Gnetum is too scarce to compare with. For example, H. Won found only 12 Cambodian collections from the compilation of herbarium collections of Gnetum in A, BK, BKF, BM, BO, CMU, F, IBSC, K, KEP, KLU, KUN, L, M, MO, NY, P, QBG, SAN, SING, U, and US during his herbarium works between 2001 and 2002, while there are 112 collection records for Indochina excluding Thailand. Also, there were only a few taxonomic studies done for Indochinese Gnetum, such as Phengklai (1975) and Hiêp and Vidal (1996), but without thorough examination of the whole Gnetum species in the Asia. Without considerate understanding of species in Asia, it can easily lead to misidentification, due to their overall similarities.
So far, we have made 77 Gnetum collections from Cambodia and 53 out of them have been deposited at the herbarium of National Institute of Biological Resources (KB). Some of them were collected without seeds or reproductive structures, which left us with uncertain identification. Also presence of putative new species without prior collection record and taxonomic review kept us from applying proper name for them. Thus, we have applied part of cp matK gene as a barcoding marker for Gnetum and tried to identify the Cambodian Gnetum collections based on phylogenetic framework.
Materials and Methods
Materials
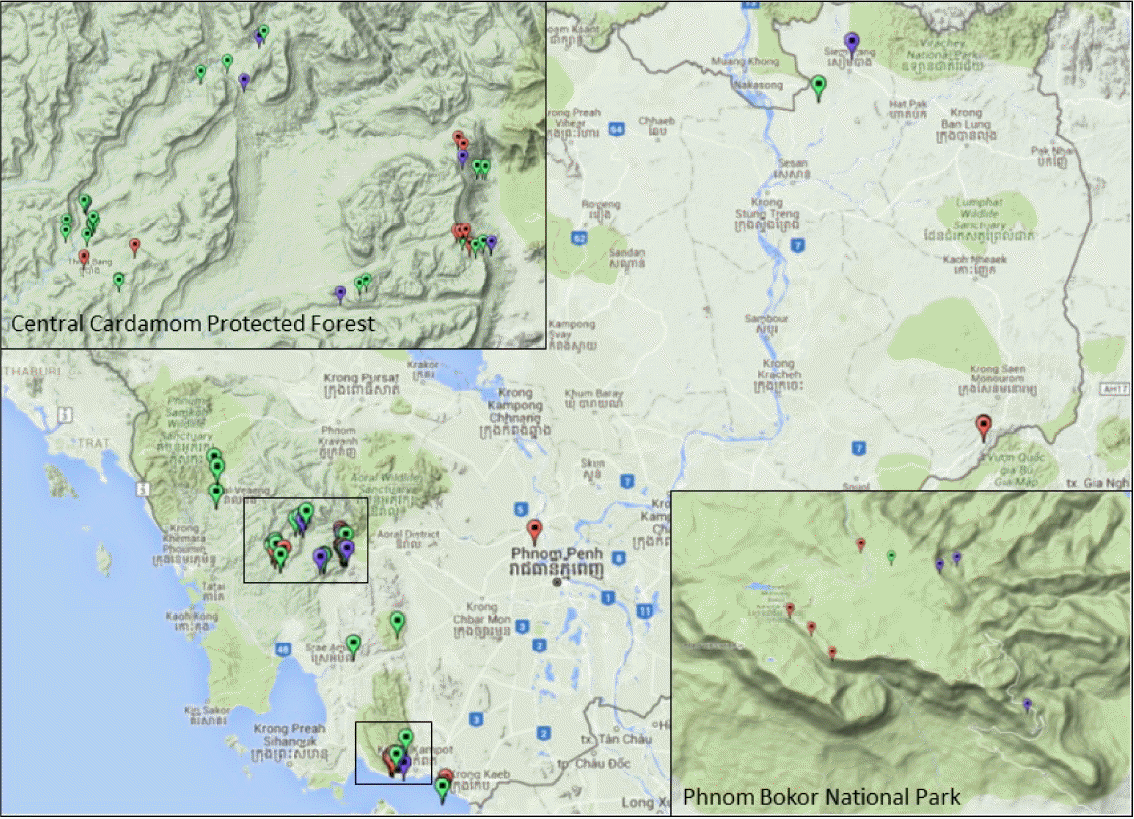
We have made 77 Gnetum collections from Cambodia between June 2010 and Nov 2015, mainly from Central Cardamom Protected Forest (Koh Kong and Kampong Speu Provinces), Phnom Bokor National Park (Kampot Province), Siem Pang (Stung Treng Province), and Seima Biodiversity Conservation Area (Mondolkiri Province), and a few from Pursat, Kandal, and Siem Reap Provinces (Fig. 1). Not all of the collections are with reproductive structure to identify confidently, and thus 53 out of 77 collections were deposited at the Herbarium of the National Institute of Biological Resources (KB), Ministry of Environment, South Korea, while all the vouchers are at the Herbarium of Daegu University (DGU). Considering the geographic distribution and availability of DNA tissue samples, we have selected 43 samples for DNA barcoding (See appendix). The exact geographic location of the 77 Gnetum specimens collected was presented over Google Map by converting GPS coordinates and collection information into KML files following the direction of Google (https://developers.google.com/kml/documentation/), and displayed over Google’s topographic map as in Fig. 1.
DNA barcode sequencing
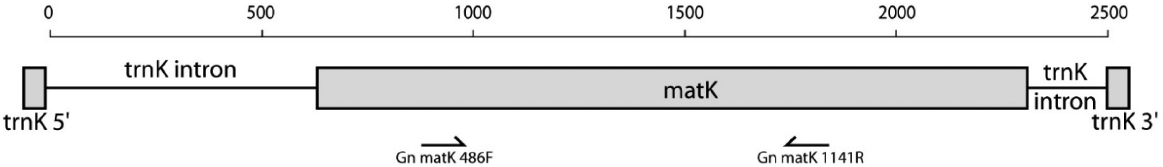
DNA extraction and sequencing followed the methods described in Won and Renner (2006) and Won (2009). For the amplification of cp matK gene region as a DNA barcode, we have applied Gn_matK_486F and Gn_matK_1141R primer combination developed by Won and Renner (2006), which amplifies base positions approximately from 486 to 1141 of cp matK gene (Fig. 2). Resulting PCR product after PCR amplification was verified under the UV lamp with known DNA size ladder, and then directly sequenced by Macrogen Inc. (Seoul, Korea), by applying the same primers for sequencing.
Analysis of DNA barcode sequences
Obtained sequence chromatograms were checked and contiged in Sequencher (ver. 4.9; Gene Codes, Ann Arbor, MI, USA; http://www.genecoes.com) and aligned in AliView (ver. 1.18; Larsson, 2014). In addition to the two representative sequences of Welwitschia and Ephedra as outgroup, 43 Gnetum matK sequences available from NCBI GenBank - 11 by Won and Renner (2006) and 29 by Hou et al. (2015) - were added to the data set of 72 newly sequenced Gnetum cp matK barcode sequences, in total making 115 Gnetum + 2 outgroup sequence data set. The data set was analyzed with PAUP* 4.0a147 (Swofford, 2002) by neighbor-joining and Maximum Parsimony criteria. The missing sites in the 5′ and 3′ ends of the sequences were trimmed out from the data matrix. Neighbor-joining tree was obtained by applying Kimura-2-parameter (K-2-P) model (Kimura, 1980). Maximum parsimony was employed for bootstrap support, by applying heuristic searches that used 100 random taxon-addition replicates, holding 100 trees at each step, tree bisection-reconnection (TBR) branch swapping, and closest taxon addition options. Nonparametric bootstrap supports were obtained by re-sampling the data 1000 times with the same search options and model for both neighbor-joining and maximum parsimony.
Results
PCR amplification and sequencing of the Gnetum cp matK barcode marker
The PCR amplification of the Gnetum matK barcode region with the Gn-matK_486F and Gn_matK_1141R primer combination was 96% successful, with only 3 out of 75 samples we have failed in amplification. However, those 3 samples have already caused difficulties in amplification of other chloroplast and nuclear marker regions with Won and Renner (2003, 2005a, 2005b, 2006). The amplified PCR products resulted in 640–714 bp long sequence contigs, with both forward and reverse sequences fully overlapping, except when the mononucleotide repeat motifs, such as A10, T8, A5GA6, A9, and T10 between the base position ∼700 and ∼850 of matK gene, sometimes presented difficulties in verifying sequences in both directions.
Phylogenetic analyses of the Gnetum cp matK barcode sequences
The neighbor-joining tree obtained from the phylogenetic analysis was presented in Fig. 3. Alignment of the obtained matK sequences was straight forward, as they are coding gene sequences. The aligned and trimmed data matrix of Gnetum matK barcode sequences were consistently 618 bp long, except two accessions, G. globosum (Chan FRI19894) and G. aff. vinkiella (Takeuchi et al. 7049), are 609 bp long. The difference is caused by the shared 9 bp long deletion (GTGTTTTTT) in the two accessions, corresponding to 3 amino acids (Val-Phe-Phe). The two accessions formed a distinct and separate clade in the phylogenetic trees obtained from neighbor-joining and maximum parsimony analyses (Fig. 3). G+C contents ranged from 28.2 to 31.2%, slightly lower than Won and Renner (2006) have reported for full matK gene sequences (31.1–33.1%), but fitting well what reported for vascular plants in general.
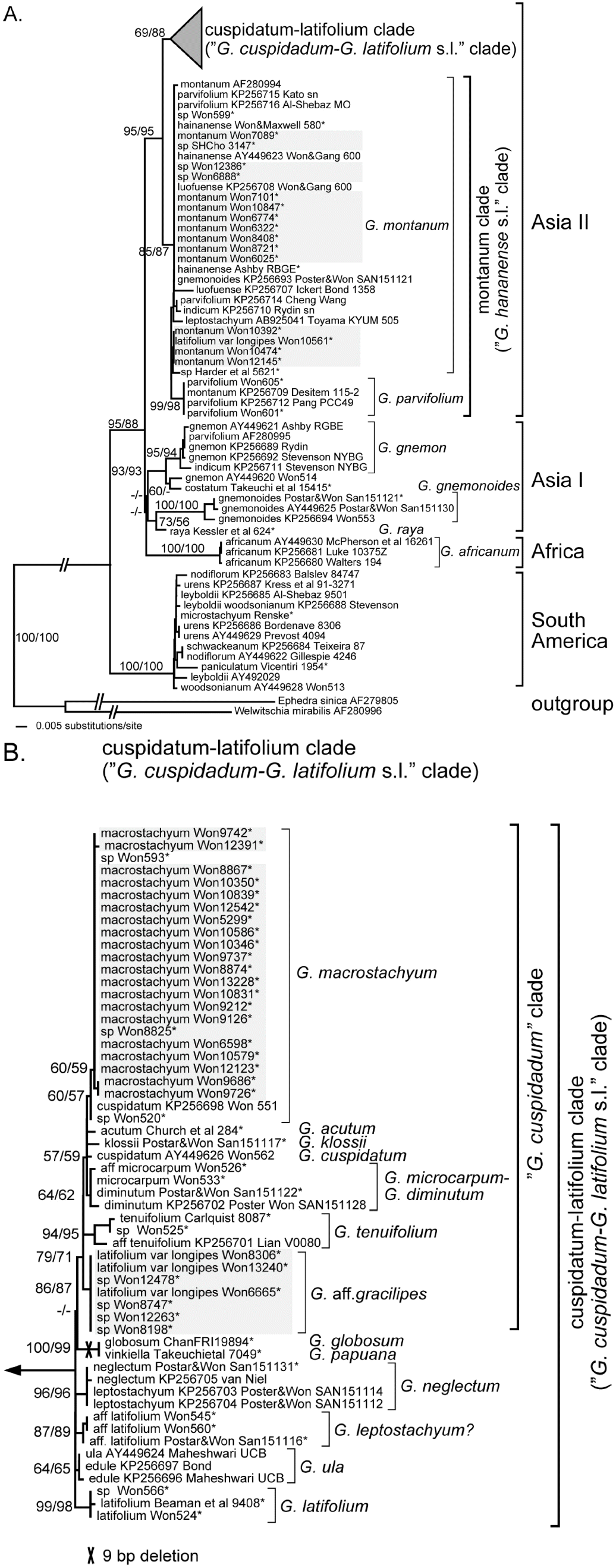
As previously presented by Won and Renner (2003, 2005a, 2005b, 2006), the current phylogenetic analyses also clustered Gnetum species into the regional clades – South America, Africa, and Asia. The monophyly of South America, Africa, and Asia-Africa clades were supported with 100–88% bootstrap supports. However, monophyly of the Asia and the Asia I clade was not supported and the sister relationship of Africa with Asia was not, either. Within the Asia I clade, monophyly of G. gnemon-G. costatum clade and that of G. gnemonoides-G. raya clade was supported with fairly strong bootstrap supports, respectively. The Asian II clade was supported with 95/95% bootstrap support, further divided into monophyletic montanum clade (formerly “G. hainanense s.l.” clade) and cuspidatum-latifolium clade (formerly “G. latifolium s.l.”, “G. cuspidatum” clades with G. ula).
Montanum clade is supported with 85/87% bootstrap support, while cuspidatum-latifolium clade with 69/88%, respectively. Within montanum clade, four G. parvifolium accessions formed a clade with 99/98% bootstrap support, distinct from other accessions named as G. montanum, G. hainanense, G. luofuense, G. indicum, and G. parvifolium. Within the cuspidatum-latifolium clade (Fig. 3B), “G. cuspidatum” clade was recognized with bootstrap support of 79/71%, with newly discovered Cambodian species G. aff. gracilipes (=G. latifolium var. longipes) included within. This is the first time the seven Cambodian samples were included and recognized as distinct species from other Gnetum species. Within “G. cuspidatum” clade, G. tenuifolium and G. aff. gracilipes branched as sister to the rest of the “G. cuspidatum” clade species, and the monophyly of the rest was supported by 57/59% bootstrap values. Those species previously recognized as members of “G. cuspidatum”, i.e., G. macrostachyum. G. acutum, G. klossii, G. cuspidatum, G. microcarpum-G. diminutum, and G. tenuifolium were retained in the clade (Won and Renner, 2003, 2005a, 2005 b, 2006). The 21 Cambodian G. macrostachyum samples, together with one accession from Cuc Phoung National Park of Vietnam (Won593), formed a monophyletic clade with 60/59% bootstrap support. Two other Gnetum samples, G. cuspidatum Won 551 and G. sp. Won 520, formed a sister relationship with G. macrostachyum clade with bootstrap support of 60/57%. The former “G. latifolium s.l.” clade was not recognized, while samples are recognized as distinct species-level clades, such as G. latifolium, G. ula, G. leptostachyum(?), G. neglectum, and G. globosum-G. papuana, with very strong bootstrap supports, except G. ula with moderate values. As mentioned above, G. globosa and G. papuana(= G. aff. vinkiella) form a distinct clade supported by shared 9-bp deletion and 100/99 bootstrap support.
Sequence divergences calculated by applying K-2-P model are presented in Table 1. South America clade shows the least sequence divergences between the accessions with 0–1.97%, except Africa clade comprised of a single species with 0% divergence, while SE Asia I clade 0–5.89% and SE Asia II 0–3.65%. Sequence divergences between the regional clades range from 4.71% to 10.03%, biggest observed between South America and Africa clades. For a species level, sequence divergences range from 0% to 1.48%, G. montanum (1.48%; excluding KP256707, 0.98%), G. tenuifolium (1.31%), G. gnemonoides (1.15%), G. gnemon (0.98%), G. microcarpum-G. diminutum (0.49%), G. macrostachyum (0.33%), while the rest were 0.16–0%.
Identification of Cambodian Gnetum
All the Cambodian Gnetum samples were clustered into three species clades - G. montanum of montanum clade, G. macrostachyum and G. aff. gracilipes of cuspidatum-latifolium clade - of Asia II clade (Fig. 3B). Out of 43 Cambodian samples, 15 were clustered into G. montanum clade, 21 into G. macrostachyum, and 7 into G. aff. gracilipes, respectively, making approximately 3 : 2 : 1 ratio for G. macrostachyum : G. montanum : G. aff. gracilipes. Seven Cambodian G. aff. gracilipes accessions show 0% sequence divergences between them, while 15 cambodain G. montanum 0–0.16% and 21 Cambodian G. macrostachyum 0–0.32%, respectively. The three Cambodian Gnetum clades are distinct from other Gnetum species clade and obviously recognizable.
Discussion
Utility of cp matK gene sequence as DNA barcode for Gnetum
Present study has shown that the cp matK gene sequences, amplified by Gn_matK_486F and Gn_matK_1141R primer combination, have sufficient resolution power to recognize Gnetum samples at the species level. The ease of amplification, sequencing, and alignment makes the cp matK a better choice over other non-coding markers. Previous work of Won and Renner (2005b), where cp trnL intron and adjacent spacers (trnT-L and trnL-F IGSs) are sequenced and analyzed, is comparable to the current work in the phylogenetic resolution, but sequence alignment was not easy due to many indels. Although the nuclear markers such as rITS and Leafy gene second intron (Won and Renner, 2005a) also have provided equally comparable resolution to these chloroplast markers, presence of putative chimeric and pseudogenic rITS copies and necessity of cloning of low copy number Leafy gene deliver difficulties and complexities in DNA barcoding, in addition to the alignment ambiguities. However, more phylogenetic information by combining those markers will be inevitable to resolve the phylogenetic relationships of the Gnetum, as tried in Won and Renner (2006). Currently, we are applying the conserved part of nuclear ribosomal ITS region to increase phylogenetic information.
Taxonomic identity of Cambodian Gnetum
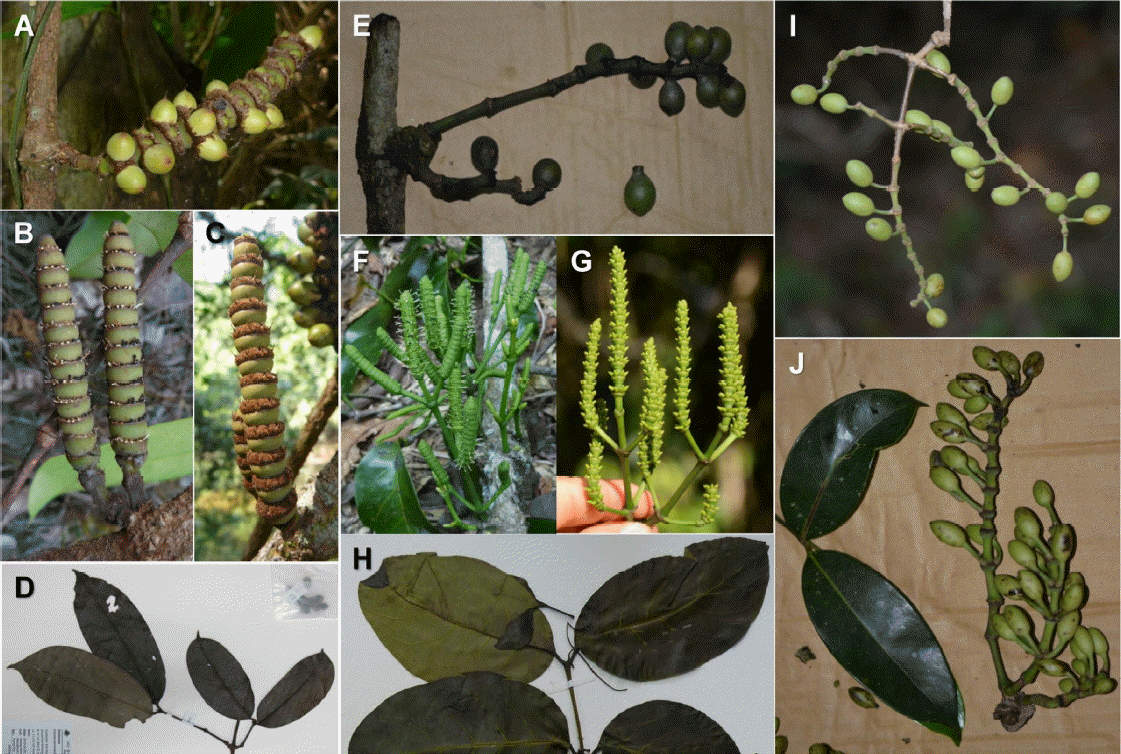
Current study identified all the Cambodian Gnetum samples into three species, G. macrostachyum, G. montanum, and G. aff. gracilipes. While G. macrostachyum has unbranched cones born on nodes, G. montanum and G. aff. gracilipes have cones born on primarily or secondarily branched rachis (inflorescence branching) at the apex of shoots or stem nodes (Fig. 4).
Gnetum macrostachyum is distributed from Indochina to Malay Peninsula, Sumatra and Borneo (Markgraf, 1929; Phengklai, 1975; Hiêp and Vidal, 1996; Won, pers. obs.). Most of the collections are from Malay Peninsular to Indochina, including Malaysia, Thailand, Vietnam, Cambodia, and Laos. Historic collections at Paris Herbarium (P) for Cambodia were made from Cardamom Mountains of Koh Kong Province and Siem Reap (Martin 99 [P!], Martin 303 [P!], Martin 330 [P!]; Hiêp and Vidal, 1996; Won, pers. obs.). We have collected 44 G. macrostachyum from Stung Treng, Siem Reap, Mondolkiri, Pursat, Kampong Speu, Koh Kong, Kampot, and Kep provinces, elevations ranging from 80 m to 940 m (Fig. 1). Current study has clearly identified G. macrostachyum distinct from other Gnetum species and there are little difficulties in identifying it from field or herbarium, due to its unique morphological characteristics. Gnetum macrostachyum is the most commonly encountered species in Cambodia and easily distinguished from other Cambodian species by its leaves and reproductive structure (Fig. 4). Leaves are oblong, abruptly narrowed to acuminate apex, base cuneate to round, leathery, thick, flat and shining. Male and female cones are born on nodes of stem, with long, abundant brown hairs. Cones are thicker than 6 mm and longer than 6 cm and not branching. Seeds are ovoid, 1–1.5 cm long with spiny tips. It shows widest habitat ranges from shaded, humid forest to partly open, dry area.
Gnetum montanum needs further taxonomic revision and comparative studies including species of southern China (Cheng et al., 1975; Shao, 1994; Fu et al., 1999a, b) and Indochina (Phengklai, 1975; Hiêp and Vidal, 1996). One of the major reason caused confusion and uncertainty of G. montanum is by author of the name Markgraf (1929) himself. Markgraf (1929) did not specify the type and enlisted 41 collections (treated as syntypes later; Price, 1996) which are collected from eastern India to Southern China including Sikkim, Assam, Birma (Myanmar), Tenasserim (Thailand), Siam (Thailand), Tonkin (Vietnam), Yunnan (China), and Kwangsi (China). Fu et al. (1999a) pointed out that the type materials and protologues are heterogeneous since they also include materials of G. latifolium. In addition, Markgraf (1929, 1951) recognized f. megalocarpum, for larger seeded G. montanum, and f. parvifolium (Warb.) Markgraf (≡G. parvifolium; Cheng, 1964), for smaller seeded and leaved ones, respectively, parallel to the varieties of G. latifolium such as var. blumei (≡var. latifolium; Markgraf, 1951), var. brachypodum, var. funiculare, var. macropodum, var. laxifrutescens, var. minus and f. longipes (≡var. longipes [Markgraf] Hiêp). Also, Markgraf (1930) recognized a new variety for G. leptostachyum, var. elongatum, from the materials collected from Annam (Vietnam; Poilane 8077 [P!, holotype & isotype]), Laos (Poilane 15646 [BKF!, P!]), and Tonkin (Vietnam; Poilane 13168 [P!]), whose distribution overlaps with G. montanum, while the rest of the G. leptostachyum is limited to Borneo (var. tenue [≡var. leptostachyum] and var. robustum) or Philippines (var. abbreviatum; Markgraf, 1951). Thus, the materials in Indochina with branched reproductive structures have been treated as G. montanum, G. latifolium, or G. leptostachyum var. elongatum (Phengklai, 1975; Hiêp and Vidal, 1996). Likewise, Gnetum of southern China had been treated as G. montanum, G. parvifolium, or G. indicum until new species were recognized for China (Cheng et al., 1975; Shao, 1994; Fu et al., 1999a, 1999b).
Current study included almost all the samples/sequences of Won and Renner (2003, 2005a, b, 2006) and Hou et al. (2015), identified as G. montanum, G. hainanense, G. luofuense, G. indicum, or G. parvifolium. All these samples were clustered only into G. montanum or G. parvifolium of montanum clade (former “G. hainanense s.l.” clade) without further division in our study (Fig. 3). This suggests that the taxonomic confusion about Gnetum samples in the Indochina and southern China exists and the diagnostic characters need further re-evaluation and verification, preferably on the basis of DNA barcoding results. For example, the species recognized by Cheng et al. (1975), such as G. hainanense (later validated by Fu et al., 1999b), G. luofuense, and G. cleistostachyum, show overlapping variations in size of seeds and stipe with G. montanum, except for G. pendulum and G. gracilipes, both having distinct ellipsoid seeds with characteristically long and slender stipes. Also descriptions of two new species recognized by Shao (1994), G. catasphaericum and G. giganteum, overlaps with those of other Chinese species, but their identity has not been verified by Fu et al. (1999a) for the Flora of China.
All the samples of montanum clade, except those four samples clustered as G. parvifolium with 99/98% bootstrap support, are clustered as G. montanum (formerly “G. hainanense s.l.”). These samples are collected from Cambodia (15 samples + Toyama KYUM 505), northern Thailand (Chiang Mai, Won&Maxwell 580), Vietnam (Harder et al. 5621), Southern China (Won 599, Won&Gang 600), and some without specific locality information un-available but definitely from southern China (Ickert Bond 1358, Cheng Wang s.n., Kato s.n., Rydin s.n., etc.). This strongly suggests that those Gnetum samples distributed from Indochina to southern China may belong to a conspecific species, with minor morphological variations in seed and leaf. Also, those recognized as G. leptostachyum var. elongatum may also belong to the G. montanum. As the other G. leptostachyum varieties are limited to the islands of Borneo and Philippines, it’s unlikely that G. leptostachyum occurs in Indochina due to geographic separation. Especially, overall morphology of the type materials matches well with those of G. montanum clade, not with other G. leptostachyum (H. Won, pers. obs.). Two accessions, Won&Maxwell 580 and Toyama KYUM 505 were originally identified as G. leptostachyum var. elongatum, but all resolved as G. montanum clade member (Fig. 3).
Overall, G. montanum shows the widest distribution from eastern India to Southern China, elevations from sea level to 3,000 m. We have collected 23 G. montanum from Mondolkiri, Kandal, Kampong Speu, Koh Kong, Kampot, and Kep Provinces of Cambodia, with elevations ranging from 50 m to 1013 m. Gnetum montanum is identified with its broad oblong-ovate leaves with more or less undulating lamina, base widely rounded, cones(+rachis) usually at shoot apex or stem nodes, cones born on primarily or secondarily branching rachis, seeds with short (3–5 mm) stipes at base or not, seeds ovoid to oblong, ∼2.5 cm long (Fig. 4).
For G. latifolium, Markgraf (1929, 1951) suggested the large oval to elliptic leaves, seeds with slender and elongated stipe, and secondarily branching cone rachis as diagnostic characters and recognized several varieties in it. Phengklai (1975) and Hiêp and Vidal (1996) recognized G. latifolium in the flora, and Markgraf (1929, 1951) also recognized a few collections from Indochina while most of the collections are limited north to the peninsula part of Malaysia. Current molecular phylogenetic analysis, consistent with previous works, has recognized a distinct clade for G. latifolium collected from Malay Peninsula (Won524, Won566) and Borneo (Beaman et al. 9408), and placed almost all the materials from northern Thailand and Indochina in G. montanum clade. This is in accord with what Won and Renner (2006) suggested for the disjunct geographic distribution pattern between clades including G. latifolium and G. montanum.
The third Cambodian Gnetum species is characterized by its ellipsoid seeds with long slender stipes and leaves elliptic to narrowly oblong (Fig. 4). We have made only three collections complete with seeds from Cardamom Ranges (Koh Kong and Kampong Speu Provinces) and Phnom Bokor National Park (Kampot Province) (Fig. 1). Five more collections were only with vegetative parts, but four out of the five are confirmed as the third species by DNA barcoding (Fig. 3). Compared to the two other Cambodian species, this occurs in the shaded humid forests, elevations from 80 to 800 m above sea level of Stung Treng, Koh Kong, Kampong Speu, and Kampot. The overall seed morphology of the third Cambodian Gnetum species fits well with that of G. latifolium var. longipes recognized by Hiêp and Vidal (1996) and Fu et al. (1999a). Gnetum latifolium var. longipes is characterized by its ellipsoid seed with long slender stipe and distributed in Philippines, Celebes, New Guinea, Solomon Islands, and Indochina (Markgraf, 1929, 1951). However, considering its disjunct distribution from other G. latifolium and narrower leaves, the Cambodian materials may represent different lineages from G. latifolium, and because of this, we are suggesting the third species need to be named independent of G. latifolium, as G. gracilipes, as suggested by Fu et al. (1999a). Further research with relevant Chinese materials will corroborate the identity and phylogenetic position of the newly found Cambodian G. gracilipes. Putative G. gracilipes specimens can be available from northern Thailand, Vietnam, and southern China, which has been overlooked so far.
One of the G. gracilipes sample with typical seeds (Won10561), though, resulted in the G. montanum clade by DNA barcoding (Fig. 3A). As Gnetum species are woody climbers with leaves on canopy while seeds born at nodes under the canopy, there is high possibility that we collected reproductive structure of G. gracilipes with vegetative tissues of G. montanum, or caused by contamination of DNA material in handling. Re-extraction of DNA from the reproductive tissues will clarify the problem.
Re-evaluation of Gnetum Samples by DNA barcoding
Based on the DNA barcoding results, there are several accessions in discordant with taxonomic identification. Reevaluation of the voucher specimens usually revealed misidentification mainly due to the lack of reproductive material to confirm identification, or by simple lack of taxonomic knowledge. For example, two accessions sequenced and analyzed by Hou et al. (2015) in the G. neglectum clade, Postar&Won San151112 and San151114, were preliminary identified as G. leptostachyum at the time of collection, but later re-identified as G. neglectum by H. Won. Hou et al. (2015) seem to simply have followed the label information without proper identification of the material. Three accessions in the G. leptostachyum(?) clade are only vegetative samples, and thus their identities have been in question. One accession, G. cuspidatum Won 551, in the G. macrostachyum clade was collected from the living collection of the Bogor Botanical Garden, Java, Indonesia, but lack of reproductive tissue may have caused the misidentification. The G. indicum accession, KP256711 from New York Botanical Garden, also suggests mis-identification of the material. The accession is clustered in the G. gnemon clade, not in any of the woody climber clades. As G. gnemon is one of the only two tree species of Gnetum, it is unlikely to mistake the samples if the authors have seen it. There are two samples sequenced and analyzed both by Hou et al. (2015) and this study, such as G. gnemonoides Postar&Won San151121 and G. montanum Won&Gang 600, but with different results. The accession of G. gnemonoides KP256693 clustered in G. montanum clade, while current study placed it in G. gnemonoides clade. To the contrary, the sequences of G. montanum Won&Gang 600 were identical. These results strongly suggest that identification by DNA barcoding can only be reliable and trusted when complemented by good taxonomic identification.