Many endangered species have small population sizes, sometimes, in extreme situations, with less than 10 remaining individuals (Li et al., 2012; Choi et al., 2013). Understanding the genetic and ecological characteristics of endangered plants or populations is important for the examination of the origin and conservation value (Isagi and Kaneko, 2014; Kaneko et al., 2017; Shiga et al., 2017) and development of effective conservation management programs (Izuno et al., 2012; Choi et al., 2013; Kaneko et al., 2013). The consequences of a small population size have received considerable research attention in the past decades (Ouborg et al., 2006; Li et al., 2012; Choi et al., 2013; Kaneko et al., 2013). In small populations, genetic drift and inbreeding can play essential roles in altering genetic variation, which can in turn influence persistence (Schaal et al., 1991; Neel and Ellstrand, 2001; Li et al., 2012). Both genetic drift and inbreeding represent potential threats for the persistence of small populations, as these processes can cause reduction of within-population genetic diversity (Ouborg et al., 2006; Willi et al., 2006) and inbreeding depression (Keller and Waller, 2002), respectively. Therefore, a precise evaluation of the current genetic status of critically endangered species or population is a necessary condition for successfully preventing their extinction (Bernardos et al. 2006).
In order to maximize the probability of success and to minimize the risk of failure of the conservation programs for critically endangered species, it is important to perform genotyping for all individuals of the target species using microsatellite markers (Izuno et al. 2012; Choi et al., 2013; Kaneko et al., 2013, 2017; Shiga et al., 2017). Analysis of the resulting genotypic data is likely to provide valuable information, such as the exact amount of genetic diversity in both wild and ex situ populations, the kinship among remnant individuals, and the extent of inbreeding and outbreeding (Choi et al., 2013; Kaneko et al., 2013, 2017). This information also can help conservation managers to identify individuals that are of particular importance to maintain and recover the genetic diversity of the target species (Choi et al., 2013).
Hydrangea luteovenosa Koidz. (Hydrangeaceae Dumort.) is a deciduous shrub that grows on the forest floor of temperate forest in Jeju Island of Korea and western Japan (Kim, 2009). Although H. luteovenosa is common in Japan, it is listed as a critically endangered plant species in Korea at regional level based on the criteria of International Union for Conservation of Nature (Kim, 2009) with a noticeably small size population (Ito et al., 2013). Only one population has been known in Seongneol-oreum region of Mt. Hallasan in Jeju Island (Moon et al., 2004; Ito et al., 2013). In conservation of endangered species or population with limited distribution like H. luteovenosa in Korea, it is important to understand the ecological status such as its breeding and clonal distribution. Recently, Ito et al. (2013) developed microsatellite markers for this species in order to investigate its genetic status. Therefore, the main purpose of this research was to evaluate the population and genetic status of the critically endangered H. luteovenosa in Korea by complete genotyping of all known individuals using nuclear microsatellite markers.
Materials and Methods
Census of the remnant population and sample collection
We initially reviewed the data on the detailed distribution of H. luteovenosa from the recently available literature (Moon et al., 2004; Kim, 2009). From the result, we visited the only known locality on Jeju Island where H. luteovenosa had previously been reported, during the period July 14 to 15 and September 25,2011. We also surveyed other areas considered suitable for this species in Seongneol-oreum region of Hallasan National Park. All wild individuals were mapped and sampled. To obtain more precise information on population structure, we examined pairwise geographic distance among individuals within the population. In addition, we visited three populations of Hyogo, Kagoshima, and Yamaguchi Prefectures in Japan, and collected leaf samples (43, 45, and 25 samples, respectively) (Fig. 1).
DNA extraction, microsatellite genotyping, and analysis
Total genomic DNA was extracted from approximately 10mg silica gel-dried leaf materials using a modified CTAB method (Milligan, 1992). All plant individuals were genotyped using five microsatellite loci recently developed for this species (Ito et al., 2013). Polymerase chain reaction (PCR) amplification mixtures were prepared following the standard protocol of the Qiagen Multiplex PCR Kit (Qiagen, Hilden, Germany) in a final volume of 10 µ L, which contained 5 ng of extracted DNA, 5 µL of 2× Multiplex PCR Master Mix, and 0.2 µM of each multiplexed primer. PCR amplifications were performed with the GeneAmp PCR System 2700 thermal cycler (Applied Biosystems, Foster City, CA, USA) using the following conditions: initial denaturation at 95oC for 15 min, followed by 30 cycles of denaturation at 94oC for 30 s, annealing at 54oC for 1 min 30 s, extension at 72oC for 1 min, and final extension at 60oC for 30 min. The size of the PCR products was measured using a ABI PRISM 3100 Genetic Analyzer (Applied Biosystems) and GENOTYPER analysis software (Applied Biosystems).
The number of alleles per locus (A), observed heterozygosity (HO), and expected heterozygosity (HE) were calculated to quantify genetic variation and connectivity in the Korean and Japanese populations. The significant deviation from Hardy-Weinberg equilibrium over all loci was tested by randomization using the FSTAT software (Goudet, 1995). Significance levels were adjusted using Bonferroni correction for multiple tests. Principal coordinate analysis (PCO) was also performed to evaluate genetic relationships among genotypes detected from Korean and Japanese populations using GenAlEx ver. 6.5(Peakall and Smouse, 2012).
Results and Discussion
Population status
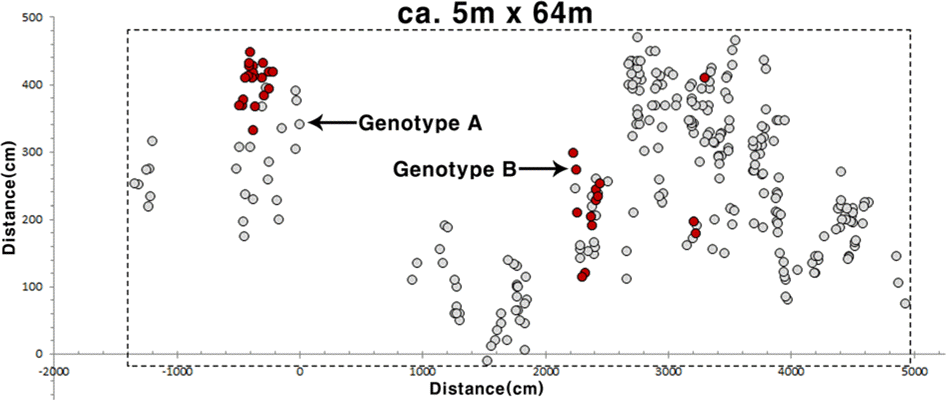
The only wild population we recorded in this study occurred in the only previously known site (Standard coordinate:33 o22′017″N, 126 o38′027″E) in Jeju Island; we did not record any new locations for Korean H. leuteovenosa. The only Korean population located in the broad-leaved deciduous forest valley dominated by Quercus serrata Murray, Carpinus laxiflora (Siebold & Zucc.) Blume, and Sasa palmata (Bean) E. G. Camus. Currently, the main threats to the wild population of Korean H. luteovenosa are (1) its small population size, consisting of 285 individuals (distribution area is ca. 5m × 64m)(Fig. 2), and (2) relatively small physical sizes of plants compared with plants in the larger Japanese population (Fig. 3). This small biological fitness can be affected by its relatively younger ages of plants compared to the Japanese population. However, it also might be influenced by negative genetic effects on population quality or inappropriate habitat environment.
Genetic status
Detailed genotyping was performed using five microsatellite markers for the all extant plants in Korea, and three Japanese populations were also genotyped for the comparative analyses (Table 1). Among the five microsatellite loci in Korean H. luteovenosa three showed polymorphism, and the number of alleles per locus (A) is 1 or 2 (Table 1). The Jeju population of H. luteovenosa showed remarkably lower genetic diversity and number of genotypes than the Japanese populations, and did not have any specific allele in the five microsatellite loci (Table 1). The 285 samples collected from the Jeju population showed only two multilocus genotypes at the five loci (Table 1), and the dominating (ca. 88%) genotype comprise by 251 individuals (Genotype A in Fig. 2). PCO analysis showed an overlapping pattern of genetic relationship among the Japanese and Jeju genotypes (Fig. 4). This result suggests that the two genotypes observed from Jeju Island have no genetic specificity in terms of their alleles. More or less disjunct distribution of individuals in each genotype (Fig. 2) might be influenced by irregular environmental features including valley overflow during the rainy season together with physical obstacles such as rocks and a forest road crossing the habitat (H. J. Choi et al., pers. obs.)
Table 1.
Variability of five microsatellite loci in four population of Hydrangea luteovenosa in Korea and Japan.
| Loci |
Jeju (Korea) (n = 285, G = 2) |
Hyogo (Japan) (n = 43, G = 42) |
Kagoshima (Japan) (n = 45, G = 45) |
Yamaguchi (Japan) (n = 25, G = 24) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | Genotype | HO | HE | A | HO | HE | A | HO | HE | A | HO | HE | |
| Hlut157 | 2 | (219, 225) | 1.00a | 0.50 | 13 | 0.81 | 0.86 | 18 | 0.84 | 0.92 | 15 | 0.81 | 0.83 |
| Hlut170 | 1 | (151, 151) | 0.00a | 0.00 | × | − | − | × | − | − | × | − | − |
| Hlut214 | 2 | (146, 150) | 1.00a | 0.50 | 9 | 0.45* | 0.8 | 6 | 0.47* | 0.52 | 2 | 0.08* | 0.18 |
| Hlut226 | 1 | (172, 172) | 0.00a | 0.00 | 3 | 0.45 | 0.43 | 2 | 0.44 | 0.47 | 2 | 0.04 | 0.04 |
| Hlut234 | 2 |
A (219, 221), B (221, 221) |
0.93a | 0.50 | 15 | 0.48* | 0.9 | 21 | 0.71 | 0.76 | 11 | 0.73 | 0.83 |
Fig. 4.
Principal coordinate (PCO) analysis of 113 genotypes in Jeju and three Japanese populations.

From the result we can conclude, (1) the Jeju population having 285 individuals consisted of only two multilocus genotypes, including identical heterozygous genotypes at two loci indicating without the recombination of sexual reproduction, (2) this predominance of only two genotypes was not observed from native Japanese populations, and (3) although the Jeju population is geographically far from Japanese populations, all alleles observed in the Korean population were shared with Japanese populations, suggesting the possibility that H. luteovenosa in the Jeju Island had been recently migrated or introduced from Japan.
Unfortunately, it is difficult to determine exact origin of Korean H. luteovenosa. There is no appropriate approach that can examine the man-introduced origin hypothesis based on present study. However, a genetic analysis about “Endangered” species suspected of man-introduced origin also reported the quite low genetic variation (Isagi and Kaneko, 2014; Kaneko et al., 2017). The results of these previous studies are similar in the current genetic status of Korean H. luteovenosa population and might support man-introduced origin hypothesis.
Implication for conservation
The Korean H. luteovenosa is considered as critically endangered plant species in Korea at regional level (Kim, 2009) with a noticeably small size population (Ito et al., 2013). Our results showed remarkably low genetic diversity especially by vegetative reproduction in the Korean population (Table 1).
However, the present findings might also demand a reconsideration of the conservation status and especially origin of Korean H. luteovenosa. Future ecological and genetic studies associated with negative effects of low genetic variation will be essential for determining the conservation direction of the threatened Korean population of this species.