적 요홍도고들빼기는 이전 연구에서 형태적 특성과 지리적 분포를 바탕으로 이고들빼기와 갯고들빼기의 잡종인 Crepidiastrum ×muratagenii로 제안된 바 있지만, 이에 대한 분자적 증거를 제시하지 못하였다. 본 연 구는 홍도고들빼기의 잡종 기원을 밝히기 위하여 홍도고들빼기와 그 근연종의 추가적인 형태적 형질을 관찰 하였으며, 핵리보솜 internal transcribed spacer (ITS) 구간과 엽록체 구간(trnT-L, trnL-F, rpl16 intron, rps16 intron)의 염기서열을 비교·분석하였다. 형태적 특성을 검토한 결과, 잡종형은 생육형, 줄기잎, 외총포편, 수과 의 특성을 바탕으로 세 가지 유형으로 구분되었다. 분자 분석 결과, Type 1형과 Type 2형은 ITS 구간의 종식 별부위에서 혼성화가 관찰되었으며, 엽록체 구간에서는 Type 1형은 이고들빼기, Type 2형은 갯고들빼기 서 열이 각각 관찰되었다. Type 3형은 ITS와 엽록체 구간 모두 이고들빼기와 동일한 서열을 보였다. Type 1형과 Type 2형은 이고들빼기와 갯고들빼기의 형태가 혼합되어 나타날 뿐 아니라, 분자 분석에서도 절영풀이 아닌, 갯고들빼기와 이고들빼기의 종식별부위에서 혼성화가 관찰되어 이고들빼기와 갯고들빼기의 잡종임을 지지 하였다. 그러나 Type 3형은 형태적 형질이 다른 잡종형과 유사하나 외총포편이 이고들빼기와 유사한 점에서 구분되며, 분자 분석에서도 이고들빼기 서열과 동일하여, 이고들빼기의 생태변이로 판단되었다.
AbstractThe plant “Hong-do-go-deul-ppae-gi” has been considered as Crepidiastrum × muratagenii, a hybrid between C. denticulatum and C. lanceolatum, based on its morphological traits and geographical distribution. To reveal the hybrid origin of Hong-do-go-deul-ppae-gi, we examined additional morphological traits of this plant and its putative parents (C. denticulatum, C. lanceolatum, C. platyphyllum) and analyzed one nuclear ribosomal internal transcribed spacer (ITS) region and four chloroplast regions (trnT-L, trnL-F, rpl16 intron, and rps16 intron). As a result of examining the morphological traits, putative hybrid individuals were classified into three types based on the habit, cauline leaf, outer phyllary, and achene beak traits. A molecular analysis found that the ITS sequences of Type 1 and Type 2 individuals showed additive species-specific sites of C. denticulatum and C. lanceolatum. Plastid sequences of Type 1 and Type 2 individuals showed C. denticulatum and C. lanceolatum sequences, respectively. However, Type 3 individuals had ITS and plastid sequences corresponding to C. denticulatum. Accordingly, Type 1 and Type 2 individuals not only share morphological traits with C. denticulatum and C. lanceolatum but also show additive species-specific sites for C. denticulatum and C. lanceolatum, and not C. platyphyllum, supporting its origin as a hybrid between C. denticulatum and C. lanceolatum. Type 3 had morphological traits similar to other hybrid types but was distinguished with respect to outer phyllaries and demonstrated some resemblance to C. denticulatum. In a molecular analysis, Type 3 was found to be identical with regard to the sequence of C. denticulatum and was judged to be an ecological variation of C. denticulatum.
INTRODUCTIONThe genus Crepidiastrum Nakai belongs to the Asteraceae (Compositae) family, with approximately 15 species distributed from Central to Eastern Asia (Shih and Kilian, 2011). The genus Crepidiastrum has been divided into three sections; section Crepidiastrum, section Monostemma Nakai, and section Paraixeris (Nakai) Pak & Kawano, based on the habit, inflorescence position, number of inner phyllaries and florets, achenes shape (Pak and Kawano, 1992).
Among the taxa in the genus Crepidiastrum, C. ×muratagenii H. Ohashi & K. Ohashi, C. ×nakaii H. Ohashi & K. Ohashi, C. ×semiauriculatum N. Yamam. & H. Ikeda, C. ×surugense (Hisauti) Yonek. have been reported as hybrid taxa (Ohashi and Ohashi, 2007; Yamamoto et al., 2009). Such hybrid taxa make it difficult to understand the taxonomic boundaries among species within the three sections.
Previous studies on Crepidiastrum hybrid taxa have mainly been conducted on C. ×nakaii [= ×Crepidiastrixeris denticulato-platyphylla (Makino) Kitam. nom. inval.] based on morphological comparison, artificial hybrid examination, cytological and molecular analysis (Ono and Satô, 1935; Saito et al., 2003, 2006). However, the other hybrid taxa have not been studied.
A plant “Hong-do-go-deul-ppae-gi” has been considered C. ×nakaii [= ×Crepidiastrixeris denticulato-platyphylla (Makino) Kitam. nom. inval.] (Lee, 1969; Lee, 1996), a hybrid between C. denticulatum (in sect. Paraixeris) and C. platyphyllum (in sect. Monostemma). However, Jang and Choi (2021) suggested that the species should be considered C. ×muratagenii, a hybrid between C. denticulatum (in sect. Paraixeris) and C. lanceolatum (in sect. Crepidiastrum), based on its morphological traits and geographical distribution. However, Jang and Choi (2021) only examined individuals from Hongdo Island in based on analysis of nuclear ribosomal internal transcribed spacer regions (nrITS), and Hongdo Island individuals did not differ in sequence from C. denticulatum. Therefore, Hong-do-go-deul- ppae-gi has not yet been clearly identified taxonomically. Hybridization usually occurs in geographical locations where the parent species are sympatric. Hence, it is necessary to observe individuals in an area where C. denticulatum and C. lanceolatum co-occur. Therefore, Hongdo Island is unsuitable since only C. denticulatum is distributed in the region.
Molecular markers such as the nrITS regions have been used extensively to investigate hybrid speciation. Although in some cases, the utility of nrITS is limited in plant phylogenetic inference (Álvarez and Wendel, 2003), the molecular marker has been very useful in identifying hybrid taxa progenitors due to its biparental inheritance nature (Sang et al., 1995; Li, 2006; Du et al., 2009; Les et al., 2009; Høibová et al., 2011; Kokubugata et al., 2011). Since hybrid species originate by mixing genomes from two different species, detection of the parental genome in the putative hybrid taxa can be direct evidence of hybrid speciation. Furthermore, the addition of chloroplast DNA (cpDNA) data can help the detection and indicate the direction of hybridization in plants (Rieseberg et al., 1993; Schwarzbach and Rieseberg, 2002). Hence, it becomes necessary to examine not only internal transcribed spacer (ITS) but also cpDNA.
Additionally, previous studies (Kitamura, 1955; Saito et al., 2006; Ohashi and Ohashi, 2007) have suggested habit, margins of cauline leaves, number of involucral bracts and florets as identification keys of Crepidiastrum hybrids. However, such identification keys are not adequate, and additional morphological traits need to be reviewed.
In the present study, to reveal the hybrid origin of Hong-do-go-deul-ppae-gi, its morphological traits were observed based on specimens collected from other regions along with Hongdo Island. One nuclear ribosomal ITS region and four chloroplast regions (trnT-L, trnL-F, rpl16 intron, rps16 intron) of the putative hybrid and its closely related taxa were analyzed. Finally, the morphological traits were described.
MATERIALS AND METHODSMorphological observationTwenty-four individuals were collected from seven localities in Korea (Fig. 1). To compare the morphological traits of the putative hybrid and its closely related taxa, we examined living materials and herbarium specimens stored at the herbarium of the Korea National Arboretum (KH), National Institute of Biological Resources (KB), Inha University (IUI) and registered at the Korea National Biospecies Information System (http://www.nature.go.kr), the collection database of specimens and materials (http://db.kahaku.go.jp/webmuseum_en/). Morphological traits were observed visually and under a stereomicroscope and sizes measured using a Mitutoyo 500-196-30 Absolute Digimatic Vernier caliper (Kanagawa, Japan). Additionally, protologues, flora, and monographs were referred to.
DNA extraction and PCR amplificationLeaf material from each individual was collected in silica gel for DNA extractions with voucher specimens (deposited in the herbarium of the Korea National Arboretum, KH). Total genomic DNA was extracted from dried leaf materials with silica gel using the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The extracted DNA was electrophoresed in a 1% agarose gel to confirm the presence or absence of DNA. The concentration and quality of DNA were confirmed using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA).
Polymerase chain reaction (PCR) amplification of the ITS of the nuclear ribosomal DNA (nrDNA) and four noncoding regions (trnT-L, trnL-F, rpl16 intron, and rps16 intron) of the cpDNA was performed. The total volume of each PCR mix was 20 μL, comprising 15 μL of distilled water, 1.0 μL of each primer (50 mM), 1 unit of Taq DNA polymerase master mix (Amplicon, Rødovre, Denmark). The primers used for PCR amplification and the PCR cycle conditions are listed in Table 1. In addition, primers ITS 2 forward and ITS 3 reverse (White et al., 1990) were used as internal primers for sequencing confirmation in both directions, particularly in the hybrids that showed nucleotide polymorphisms. The PCR products were visualized in 1% agarose gels and sequenced using an ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). The determined sequences were deposited in GenBank (Table 2). Additionally, C, platyphyllum individuals deposited in GenBank were used for phylogenetic analysis. The analyzed nucleotide sequences were determined after checking the chromatogram using Geneious R 7.1.9 (Biomatters Ltd., Auckland, New Zealand). All sequences were aligned using MUSCLE (Edgar, 2004). Polymorphic sites in the ITS region were identified by overlapping peaks on the chromatogram.
Phylogenetic analysisMaximum likelihood trees based on the nrITS region and the combined four cpDNA regions (trnT-L, trnL-F, rpl16 intron, and rps16 intron) were constructed using the W-IQ-TREE Server (http://iqtree.cibiv.univie.ac.at/) (Trifinopoulos et al., 2016), based on user-friendly web servers for IQ-TREE 1.5 (Nguyen et al., 2015). Each aligned sequence dataset was tested to determine the best-fit model by using W-IQ-TREE with the Akaike criterion, and new model selection procedures. TIM3e was confirmed as the best-fit model for the selected nrITS and cpDNA regions. We evaluated the node supported by 1,000 ultrafast bootstrap replicates (UFBS) (Minh et al., 2013). Youngia japonica (L.) DC., which is most closely related to the genus Crepidiastrum, was used as an outgroup (Kilian et al., 2009).
RESULTSObservation of morphological traitsAs a result of observing the morphological traits, putative hybrid individuals were classified into three morphological types. All morphotypes had 10–12 florets but lacked basal leaves at anthesis. They grow on sunny slopes that were adjacent to mountains and coasts. However, Type 1 had herbaceous and erect stem branching in the upper part, mixed entire and serrate margins, 1.2–1.8 mm outer phyllaries, and achenes with 0.5–0.9 mm beaks. Type 2 had a suffrutescent stems with stout and short caudex, branched at the base, mixed entire and serrate margins, 1.4–1.6 mm outer phyllaries, and achenes with 0.2–0.5 mm beaks. Type 3 had a suffrutescent stem with stout and short caudex, branched at the base, mixed entire and serrate margins, 0.4–0.7 mm outer phyllaries, and achenes with 0.5–0.7 mm beaks. All three morphotypes showed that C. denticulatum and C. lanceolatum had mixed morphological traits (Fig. 2, Table 3). Type 1 and Type 2 were observed in Busan, Type 2 was observed on Somaemuldo Island, Type 2 and Type 3 were observed on Geomundo Island, and Type 3 was observed on Chujado and Hongdo Islands.
nrDNA ITS sequences and phylogenetic analysisThe sizes of the nrITS regions in C. platyphyllum, C. denticulatum, C. lanceolatum were 640 bp, 639 bp, and 638 bp, respectively. Size variation among the individuals within the same species was not observed. Four accessions of C. platyphyllum had identical sequences, while five accessions of C. denticulatum were grouped into three ribotypes (A, B, C) which differed at nucleotide position of 603, and five accessions of C. lanceolatum were grouped into three ribotypes (D, E, F) that differed at three nucleotide sites (200, 212, 434 bp) (Table 4). After alignment, a data set with a length of 641 bp was obtained. The length variation of the aligned sequence data was due to 1 to 3 bp of indels (insertion/deletion events). C. platyphyllum had 249 bp (1 indel) for the nrITS 1 region, 164 bp for the 5.8S, and 227 bp for nrITS 2 region. C. denticulatum and C. lanceolatum had 250 bp for the nrITS 1 region, 164 bp for 5.8S, but 225 bp (2 indels) and 224 bp (3 indels) for the nrITS 2 region, respectively. Excluding the gaps, the putative parental species showed sequence variation at a total of ten nucleotide sites. The pairwise-sequence difference between C. denticulatum and C lanceolatum was six nucleotide substitutions and one indel. C. denticulatum and C. platyphyllum differed by eight nucleotide substitutions and three indels. C lanceolatum and C. platyphyllum could be distinguished by six nucleotide substitutions and four indels. Therefore, the three taxa of Crepidiastrum were clearly distinguished in the nrITS regions. Among the putative hybrid types, Type 1 and Type 2 exhibited polymorphic ITS signals at those seven sites differentiating C. denticulatum and C. lanceolatum, showing sequence additivity. These were visible as double peaks in the electropherograms. Type 3 exhibited species-specific nucleotides of C. denticulatum. Additionally, no species-specific nucleotides from C. platyphyllum were observed in all putative hybrid individuals (Table 4).
The aligned nrITS sequences of the three Crepidiastrum taxa except for the hybrid putative individuals had a length of 648 characters, comprising 546 constant, 90 parsimony-uninformative characters, and 12 parsimony-informative characters. In the nrITS phylogenetic tree, three taxa of the genus Crepidiastrum were divided into two clades. C. denticulatum was formed in a weakly supported clade (bootstrap [BS] = 57). Crepidiastrum platyphyllum and C. lanceolatum were formed in a clade (BS = 94), and each taxon was formed in a monophyletic group, respectively (BS = 99, BS = 88) (Fig. 3).
Combined cpDNA sequences and phylogenetic analysisTo trace the plastid donors (maternal origin) of the putative hybrid individuals, DNA sequences of the four plastid markers (trnT-L, trnL-F, rpl16 intron, and rps16 intron) were examined for the 24 individuals representing the putative hybrid and its closely related taxa. The size ranges of the four markers observed from the examined taxa were 528 to 537 bp (trnT-L), 766 bp (trnL-F), 949 bp (rpl16 intron), 721 to 740 bp (rps16 intron), respectively. In the trnT-L, sequence length for all examined accessions was 536–537 bp except C. denticulatum, C. platyphyllum, and Type 1 which had a length of 528–530 bp due to an 8 bp deletion at nucleotide sites 4 to 11, 1 bp insertion at nucleotide site 217 and single nucleotide variation observed at nucleotide position of 18. In trnL-F, two haplotypes (A, B) were found in C. denticulatum that showed single nucleotide variation at nucleotide position of 632. In rpl16 intron, three nucleotide variations were observed at nucleotide positions of 132, 307, and 801, respectively. The rps16 intron showed two nucleotide variations at nucleotide positions of 438 and 697 and two haplotypes were found in C. lanceolatum, which differed in size (721 and 740 bp). Therefore, among the analyzed four cpDNA markers, trnT-L, rpl16 intron and rps16 intron are useful for distinguishing between C. denticulatum and C. lanceolatum. However, since C. denticulatum and C. platyphyllum differ by only a single nucleotide sequence in the rpl16 intron (at 307 bp), additional markers are required to clearly distinguish the two taxa. A plastid type of C. platyphyllum was not detected in the examined samples of all the hybrid types (Table 5).
The aligned combined chloroplast sequence (trnT-L, trnL-F, rpl16 intron, and rps16 intron) had a length of 3,036 characters, comprising 2,959 constant, 69 parsimony-uninformative characters, and 8 parsimony-informative characters. In the combined cpDNA phylogenetic tree, the putative hybrid individuals and the three taxa of the genus Crepidiastrum were divided into two clades. Crepidiastrum denticulatum, C. platyphyllum, Type 1, and Type 3 formed a strongly supported clade (BS = 80%), and C. lanceolatum and Type 2 a weakly supported clade (BS = 56%) (Fig. 4).
DISCUSSIONThe plant “Hong-do-go-deul-ppae-gi” has been regarded as C. ×nakaii, a hybrid between C. denticulatum and C. platyphyllum (Lee, 1969; Lee, 1996). However, the results of the molecular analysis of nrITS sequences of Type 1 and Type 2 individuals in the present study showed additive species-specific sites for C. denticulatum and C. lanceolatum (Table 4). This is interpreted from the results obtained by amplification of copies inherited from both parental species. Therefore, the results of the present study support the hypothesis proposed by Jang and Choi (2021), that Hong-do-go-deul-ppae-gi should be considered as C. ×muratagenii, a hybrid between C. denticulatum and C. lanceolatum. Nevertheless, Jang and Choi (2021) treated Hongdo Island individuals as a C. ×muratagenii. Type 3 included Hongdo Island individuals which had habitats and morphological traits similar to those of other hybrid types, but was distinguished by minute outer phyllaries (ca. 0.5 mm vs. ca. 1.5 mm in Type 1 and Type 2) (Fig. 2), which was identical with the sequence of C. denticulatum when the molecular analysis was performed. Hence, it was judged to be an ecological variation of C. denticulatum. Species-specific markers of C. platyphyllum were not detected in any putative hybrid individuals (Table 4). This agrees with the morphological observations, indicating that C. denticulatum and C. lanceolatum co-occurred in Busan, Somaemuldo and Geomundo Islands, and were involved in the hybridization.
The combined cpDNA sequences (trnT-L, trnL-F, rpl16 intron, and rps16 intron) of Type 1 and Type 2 showed species-specific markers for C. denticulatum and C. lanceolatum, respectively (Table 5). The results indicate that both C. denticulatum and C. lanceolatum have served as a plastid donor suggesting that, in the case of C.×muratagenii, the hybrid speciation that has occurred is bidirectional. However, no apparent geographic structuring of the haplotypes could be found. Further sampling might reveal the geographic structuring of the genus Crepidiastrum. In the combined cpDNA phylogenetic tree, C. denticulatum in sect. Paraixeris and C. platyphyllum in sect. Monostemma formed a clade (Fig. 4). However, the ITS phylogenetic tree showed that C. platyphyllum is sister to C. lanceolatum in sect. Crepidiastrum (Fig. 3). Similarly, additional sampling and markers are required to understand the relationships among the three sections and the relationships between species within a particular section.
In general, divergent ribotypes in hybrid lineages can be homogenized quickly by a mechanism known as concerted evolution (Wendel et al., 1995; Aguilar et al., 1999). However, some early generations of hybrid lineages can maintain divergent ribotypes contributed by parental species (Siripun and Schilling, 2005; Liu et al., 2009; Cho et al., 2014; Shin et al., 2014; Gil and Kim, 2016). The high number of completely additive polymorphic sites and the lack of new unique mutations in C. ×muratagenii in relation to its completely differentiated and non-sibling parental species are consistent with the expected pattern of recent or F1 hybrid formation (Rieseberg et al., 1993; Nieto Feliner et al., 2004).
The possibility of a backcross from one of the putative parents cannot be ruled out. However, we have not observed any such cases in the present molecular data, or perhaps our materials are too limited for drawing such conclusions. In other cases, regarding the C. ×nakaii from Saito et al. (2006), second or more generations have been reported and assumed. Also, the hybrid individuals observed in field research show a certain degree of fertility. There also exist a high likelihoods of backcrosses and introgressions.
The ITS and combined cpDNA (trnT-trnL, trnL-trnF, rpl16 intron, and rps16 intron) investigations provide strong evidence for the natural hybridization between C. denticulatum and C. lanceolatum. Further detailed investigations need to be performed on the pollen and ovule sterility, chromosome number, karyotype, and reproductive behavior of the hybrids.
Taxonomic treatment
Crepidiastrum ×muratagenii H. Ohashi & K. Ohashi, J. Jap. Bot. 82: 342, 2007.—TYPE: JAPAN. Kyushu: Kagoshima Pref., Yakushima, Yaku-cho, Kurio, 10 Nov 1983, G. Murata, T. Takagi & A. Iwami 40 (holotype: KYO, photo!, isotype: KYO, photo!).
= Crepidiastrum denticulatum (Houtt.) J. H. Pak & Kawano × C. lanceolatum (Houtt.) Nakai.
[×Crepidiastrixeris denticulato-lanceolata Kitam., Acta Phytotax. Geobot. 11: 132, 1942 nom.inval.]
Korean name: Hong-do-go-deul-ppae-gi (홍도고들빼기).
Herbs with erect stem and branched in the upper part or subshrubs with woody caudex stout, short, branched in the lower part, 10–30 cm tall. Leaves alternate, sessile; basal leaves withered at anthesis; blade of lower and middle cauline leaves spatulate-oblong, ovate-oblong or ovate, 4.1–7.9 × 1.5–4.2 cm, apex acute or obtuse, clasping at base, margins mixed entire and acute serrations, slightly thick texture; upper cauline leaves similar to middle leaves, 1.6–3.6 × 0.5–1.6 cm. Heads ca. 1.5 cm diam., with 10–12 florets; peduncle slender, 6.1–8.5 mm long; bracteoles many, ovate, 0.5–0.7 mm long; involucre tubular 6–8 mm long, with small bracteoles at base; outer phyllaries few, ovate, 1.2–1.8 mm long, acute or obtuse; inner phyllaries, 8, lanceolate, 5.5–7.4 mm long. Achenes fusiform, slightly compressed, brownish, 2.9–3.9 mm long, with 0.2–0.9 mm beak, 10–15 scabrid ribs; pappus 3–4 mm long, white.
Phenology: Flowering October to November and fruiting November to December
Distribution: China, Japan (Kyushu), Korea (Jeollanam-do, Gyeongsangnam-do).
Habitat: Sunny slopes adjacent to the mountains and the coast where C. denticulatum and C. lanceolatum occurred sympatrically.
Taxonomic notes: It is difficult to distinguish C. ×muratagenii because it shows mixed traits of C. denticulatum and C. lanceolatum (Fig. 2, Table 3). Crepidiastrum ×muratagenii was similar to C. lanceolatum by having outer phyllaries of 1.2–1.8 mm long (ca. 0.5 mm long in C. denticulatum), 10–12 florets (13–15 in C. denticulatum). However, it differed from C. lanceolatum by lacking basal leaves at anthesis, cauline leaves margins mixed with entire and acute serrations.
ACKNOWLEDGMENTSThe authors are grateful to the persons concerned at the KB herbaria for permitting the examination of specimens. This work was supported by the Korea National Arboretum (KNA1-1-23, 18-1).
Fig. 1.Investigated localites (JR, Mt. Jirisan; UJ, Ulju-gun; BS, Busan; SMMD, Somaemuldo Island; GMD, Geomundo Island; JJD, Jejudo Island; CJD, Chujado Island; HD, Hongdo Island). 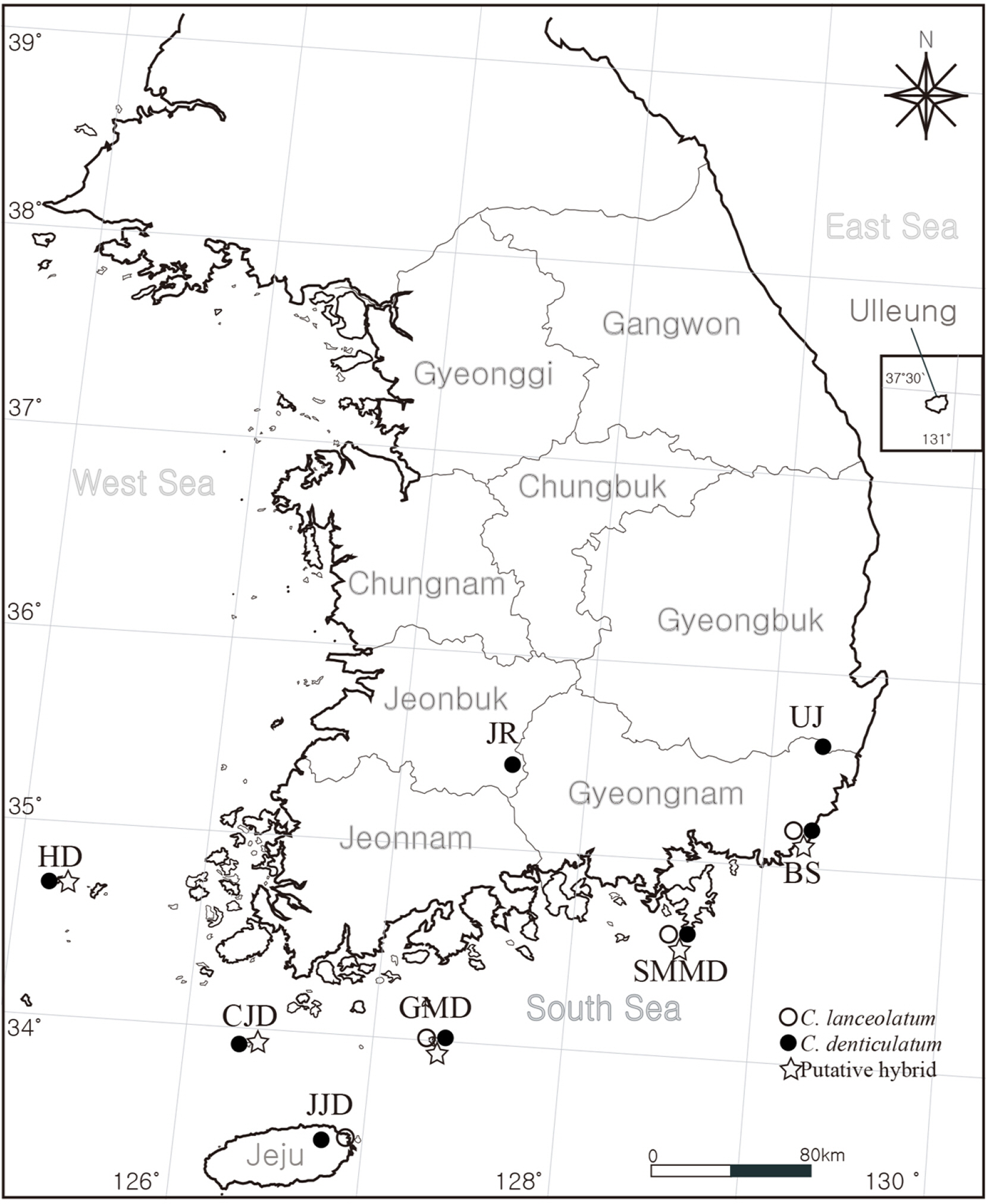
Fig. 2.Photographs of putative hybrids. 1–3. Three morphological types. A. Specimen. B. Shape of cauline leaves. C. Head, top view (l, lingule). D. Head, lateral view (i, inner phyllaries; o, outer phyllaries). E. Shape of achenes (b, beak). 
Fig. 3.Maximum likelihood (ML) tree based on the nuclear ribosomal internal transcribed spacer (nrITS) sequences of three taxa of Crepidiastrum. Bootstrap values (>50%) are indicated above branches. 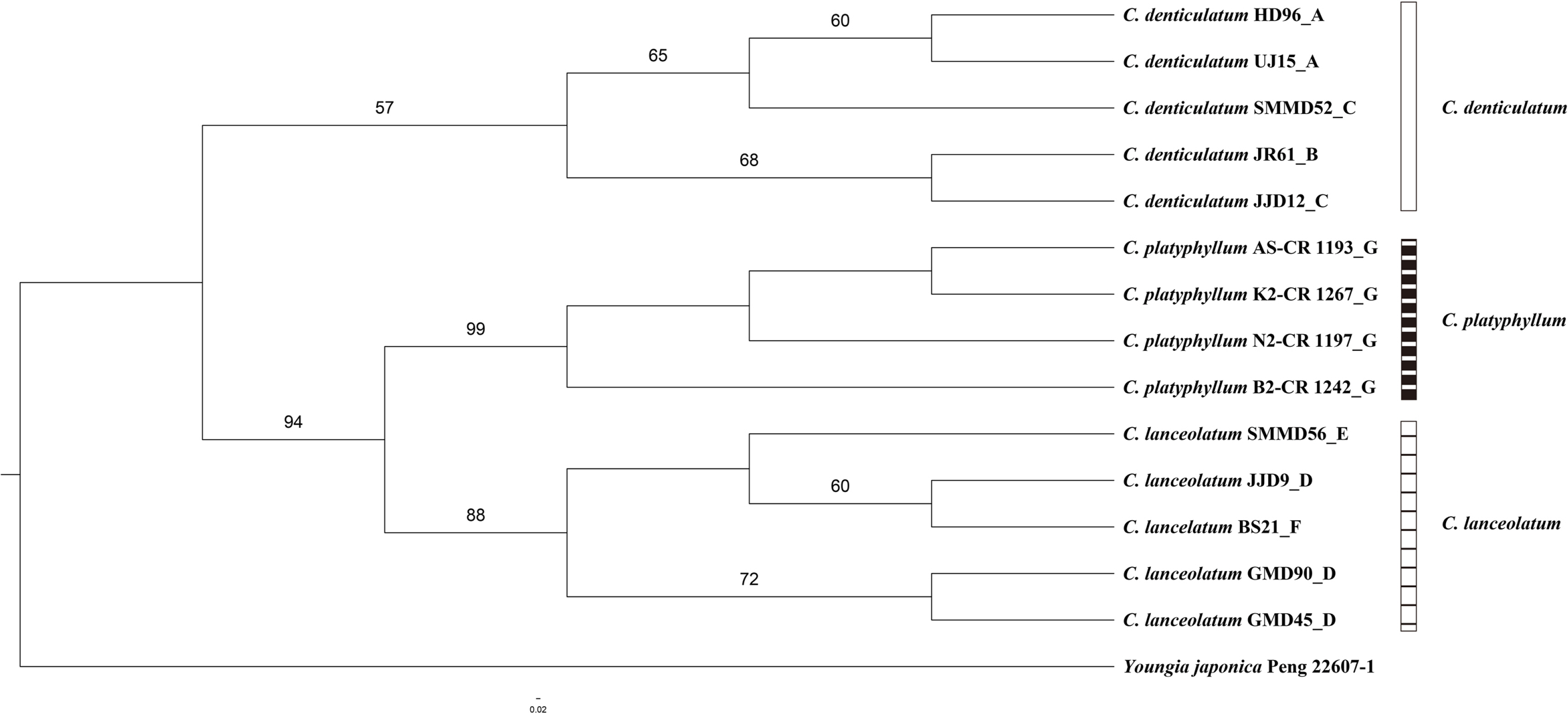
Fig. 4.Maximum likelihood (ML) tree based on the combined cp region (trnT-L + trnL-F + rpl16 intron + rps16 intron) of putative hybrid and its closely related taxa. Bootstrap values (>50%) are indicated above branches. 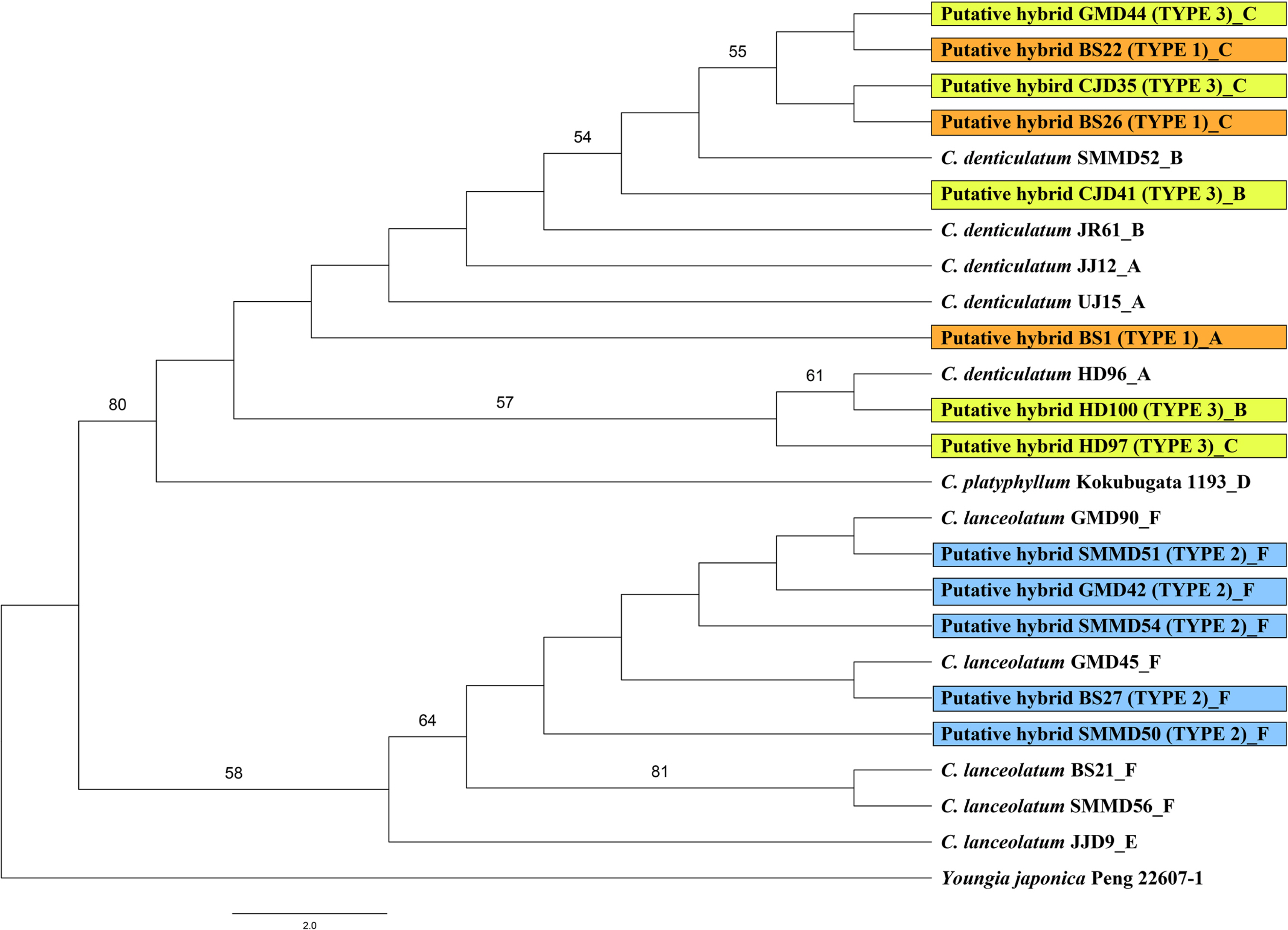
Table 1.List of primers and PCR conditions used in the present study.
Table 2.List of taxa used for nrITS and cpDNA sequences analysis with voucher and GenBank accession numbers. Table 3.Comparison of the morphological characters among putative hybrids and their closely related taxa. Table 4.Nuclear species-specific sites in putative hybrid and its closely related taxa. Table 5.Plastid species-specific sites in putative hybrid and its closely related taxa. LITERATURE CITEDAguilar, JF. Rosselló, JA and Feliner, GN. 1999. Nuclear ribosomal DNA (nrDNA) concerted evolution in natural and artificial hybrids of Armeria (Plumbaginaceae). Molecular Ecology 8: 1341-1346.
Álvarez, I and Wendel, JF. 2003. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolition 29: 417-434.
Cho, M.-S. Kim, C.-S. Kim, S.-H. Kim, TO. Heo, K.-I. Jun, J and Kim, S-C. 2014. Molecular and morphological data reveal hybrid origin of wild Prunus yedoensis (Rosaceae) from Jeju Island, Korea: Implications for the origin of the flowering cherry. American Journal of Botany 101: 1976-1986.
Du, Z.-Y. Yang, C-F. Chen, J-M and Guo, Y-H. 2009. Nuclear and chloroplast DNA sequences data support the origin of Potamogeton intortusifolius J.B. He in China as a hybrid between P. perfoliatus Linn. and P. wrightii Morong. Aquatatic Botany 91: 47-50.
Edgar, RC. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792-1797.
Gil, H.-Y and Kim, S-C. 2016.
Viola woosanensis, a recurrent spontaneous hybrid between V. ulleungdoensis and V. chaerophylloides (Violaceae) endemic to Ulleung Island, Korea. Journal of Plant Research 129: 807-822.
Høibová, E. Čížková, J. Christelová, P. Taudien, S. de Langhe, E and Doležel, J. 2011. The ITS1-5.8S-ITS2 sequence region in the Musaceae: Structure, diversity and use in molecular phylogeny. PLoS ONE 6: e17863.
Jang, Y.-J and Choi, B-H. 2021. Taxonomic identity of Crepidiastrum ×nakaii recorded in Hongdo Island. Korean Journal of Plant Taxonomy 51: 198-204.
Kilian, N. Gemeinholzer, B and Lack, HW. 2009. Cichorieae. Systematics, Evolution and Biogeography of Compositae. Funk, V. A. Susanna, A. Stuessy, TF. Bayer, RJ (eds.), International Association for Plant Taxonomy, Vienna. 343-383.
Kitamura, S. 1955. Compositae Japonicae. Pars Quarta. Memoirs of the College of Science, University of Kyoto, Series B 22: 77-126.
Kokubugata, G. Kurihara, T. Hirayama, Y and Obata, K. 2011. Molecular evidence for a natural hybrid origin of Ajuga ×mixta (Lamiaceae) using ITS sequence. Bullein of the National Museum of Nature and Science, Series B 37: 175-179.
Lee, TB. 1969. Plant resources in Korea. Bulletin of Seoul National University (Biological Agriculture) 20: 158 (in Korean).
Lee, WT. 1996. Lineamenta Florae Koreae. Academy Publishing Co, Seoul. 1688 (in Korean).
Les, DH. Murray, NM and Tippery, NP. 2009. Systematics of two imperiled pondweeds (Potamogeton vaseyi, P. gemmiparus) and taxonomic ramifications for subsection Pusilli (Potamogetonaceae). Systematic Botany 34: 643-651.
Li, W.-P. 2006. Natural hybridization between Aster ageratoides var. scaberulus and Kalimeris indica (Asteraceae): Evidence form morphology, karyotype, and ITS sequences. Botanical Studies 47: 191-197.
Liu, S-C. Lu, C-T and Wang, J-C. 2009. Reticulate hybridization of Alpinia (Zingiberaceae) in Taiwan. Journal of Plant Research 122: 305-316.
Minh, BQ. Nguyen, M. A. T and von Haeseler, A. 2013. Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30: 1188-1195.
Nguyen, L.-T. Schmidt, H. A.. von Haeseler, A and Minh, BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268-274.
Nieto Feliner, G. Gutiérrez Larena, B and Fuertes Aguilar, J. 2004. Fine-scale geographical structure, intra-individual polymorphism and recombination in nuclear ribosomal internal transcribed spacers in Armeria (Plumbaginaceae). Annals of Botany 93: 189-200.
Ohashi, H and Ohashi, K. 2007. Hybrids in Crepidiastrum (Asteraceae). Journal of Japanese Botany 82: 337-347.
Ono, H and Satô, D. 1935. Intergenera hibridigo en Cichorieae, II. Hibridoj de Crepidiastrum lanceolatum var. latifolium kaj Paraixeris denticulata. Journal of Japanese Genetics 11: 169-178 (in Esperanto).
Oxelman, B. Liden, M and Berglund, S. 1997. Chloroplast rps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae). Plant Systematic and Evolution 206: 393-410.
Pak, J-H and Kawano, S. 1992. Biosystematic studies on the genus Ixeris and its allied genera (Compositae-Lactuceae) (IV): Taxonomic treatments and nomenclature. Memoirs of the Faculty of Science, Kyoto University, Series of Biololgy 15: 29-61.
Rieseberg, LH. Ellstrand, NC and Arnold, M. 1993. What can molecular and morphological markers tell us about hybridization? Critical Reviews in Plant Sciences 12: 213-241.
Saito, Y.. Kokubugata, G. Katsuyama, T. Marubashi, W and Iwashina, T. 2003. Cytological comparisons of somatic chromosomes in ×Crepidiastrixeris denticulato-platyphylla and speculation of its parental species (Asteraceae). Chromosome Science 7: 43-48.
Saito, Y. Möller, M. Kokubugata, G. Katsuyama, T. Marubashi, W and Iwashina, T. 2006. Molecular evidence for repeated hybridization events involved in the origin of the genus ×Crepidiastrixeris (Asteraceae) using RAPDs and ITS data. Botanical Journal of the Linnean Society 151: 333-343.
Sang, T. Crawford, DJ and Stuessy, TF. 1995. Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: Implications for biogeography and concerted evolution. Proceedings of the National Academy of Sciences of the United States of America 92: 6813-6817.
Schwarzbach, AE and Rieseberg, LH. 2002. Likely multiple origin of a diploid hybrid sunflower species. Molecular Ecology 11: 1703-1715.
Shih, C and Kilian, N. 2011. Crepidiastrum. Wu, Z-Y. Raven, P H. Hong, D.-Y (eds.), Flora of China. 20–21: (Asteraceae).. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis, MO. 264-269.
Shin, H. Oh, S-H. Lim, Y. Hyun, C-W. Cho, S-H. Kim, Y-I and Kim, Y-D. 2014. Molecular evidence for hybrid origin of Aster chusanensis, an endemic species of Ulleungdo, Korea. Journal of Plant Biology 57: 174-185.
Siripun, KC and Schilling, EE. 2005. Molecular confirmation of the hybrid origin of Eupatorium godfreyanum (Asteraceae). American Journal of Botany 93: 319-325.
Small, RL. Ryburn, JA. Cronn, RC. Seelanan, T and Wendel, JF. 1998. The tortoise and the hare: Choosing between noncoding plastome and nuclear Adh sequences for phylogeny reconstruction in a recently diverged plant group. American Journal of Botany 85: 1301-1315.
Taberlet, P.. Gielly, L. Pautou, G and Bouvet, J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105-1109.
Trifinopoulos, J. Nguyen, L-T. von Haeseler, A and Minh, BQ. 2016. W-IQ-TREE: A fast online phylogenetic tool for Maximum likelihood analysis. Nucleic Acids Research 44: W232-W235.
Wendel, JF. Schnabel, A and Seelanan, T. 1995. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proceedings of the National Academy of Sciences of the United States of America 92: 280-284.
White, TJ. Bruns, T. Lee, A and Tayler, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Application. Innis, M. Gelfand, D. Sninsky, J. White, T (eds.), Academic Press, San Diego. 315-322.
Yamamoto, N. Yano, O and Ikeda, H. 2009. A new hybrid, Crepidiastrum ×semiauriculatum (Asteraceae: Lactuceae), from Okayama Prefecture, Western Japan. Journal of Japanese Botany 84: 224-228.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||