AbstractThe nucleotide sequences of the chloroplast rbcL, matK, and psbA-trnH and nuclear internal transcribed spacer (ITS) regions were determined from all species of Viburnum in Korea with multiple accessions to reconstruct the phylogeny and to evaluate the utility of the DNA sequences as DNA barcodes. The results of phylogenetic analyses of the cpDNA and ITS data are consistent with the findings of previous studies of Viburnum. Four morphologically closely related species, V. dilatatum, V. erosum, V. japonicum, and V. wrightii, were included in a strongly supported sister clade of V. koreanum and V. opulus. Viburnum odoratissimum is suggested to be sister to the V. dilatatum/V. koreanum clade in the cpDNA data, while V. odoratissimum is a sister to V. furcatum in the ITS data. Viburnum burejaeticum and V. carlesii are strongly supported as monophyletic. Our analyses of DNA barcode regions from multiple accessions of the species of Viburnum in Korea confirm that six out of ten species in Korea can be discriminated at the species level. The V. dilatatum complex can be separated from the remaining species according to molecular data, but the resolution power to differentiate a species within the complex is weak. This study suggests that regional DNA barcodes are useful for molecular species identification in the case of Viburnum when flowering or fruiting materials are not available.
The genus Viburnum L. consists of 150–200 species of small trees and shrubs widely distributed in temperate and subtropical regions of Asia, Europe, North Africa, and the Americas with high species diversity in eastern Asia and Latin America (Rehder, 1908; Donoghue, 1983; Hara, 1983; Donoghue et al., 2004). Viburnum is characterized within Adoxaceae by having simple leaves and single-stoned drupes (Donoghue et al., 2003). Plants of Viburnum have been widely utilized as ornamentals due to their showy flowers and fragrancy, such as V. opulus L., and V. carlesii Hemsl. and as a medicine source, such as V. tinus L. and V. erosum Thunb. (Mabberley, 1997; Ahn, 1998; Kwon et al., 2003).
The phylogeny of Viburnum has been extensively studied using various DNA regions in both nuclear and chloroplast genomes (Donoghue et al., 2004; Winkworth and Donoghue 2004, 2005; Clement and Donoghue 2011, 2012; Clement et al., 2014). These studies have improved our understanding of the phylogeny, the evolution of morphological characters and the biogeography of Viburnum. Most traditionally recognized sections within Viburnum (Hara, 1983) were supported as monophyletic (Donoghue et al., 2004; Winkworth and Donoghue 2004, 2005; Clement and Donoghue 2011, 2012; Clement et al., 2014). Clement et al. (2014) proposed a phylogenetic classification (Cantino et al., 2007) based on strongly supported and congruent clades among different data sets of chloroplast DNA.
In Korea, ten species of Viburnum belonging to five sections (Hara, 1983) or clades (Clement et al., 2014) are distributed (Hong and Im, 2003; Kim, 2007) (Table 1). Previous phylogenetic studies using chloroplast DNA regions attempted to resolve higher level relationships in Viburnum and included most of the species, but only one accession per species was analyzed (Clement and Donoghue 2012; Clement et al., 2014). Phylogenetic studies using nuclear genomes, such as internal transcribed spacer (ITS) regions (Donoghue et al., 2004) and GBSSI (Winkworth and Donoghue 2004), included some of the species, though the phylogenetic relationships among the species, especially those in the same clade, have remained unclear. In particular, V. burejaeticum Regel & Herder, V. wrightii Miq., and V. koreanum Nakai were not included in the phylogenetic analyses using nuclear DNA regions (Donoghue et al., 2004; Winkworth and Donoghue 2004).
The utility of DNA barcoding is well established in variety of plant taxa (Hebert et al., 2003; Kress et al., 2005; Lahaye et al., 2008; CBOL Plant Working Group, 2009; Xiang et al., 2011; Kim et al., 2014). The performance of DNA barcoding in Viburnum with rbcL, matK, rpl32-trnL, psbA-trnH, trnK, and ITS regions from 112 species showed very low levels of species resolution among closely related species. The establishment of a regional barcode database with multiple accessions encompassing the distributional range in Korea and an assessment of the levels of variation in Viburnum are necessary to make the molecular species identification tool useful (Kim et al., 2014; Youm et al., 2016).
This study aims to determine the DNA sequences of rbcL, matK, psbA-trnH, and ITS regions from multiple accessions of all species of Viburnum in Korea to assess the level of differentiation of the species and to evaluate the resolution power of DNA barcodes to identify a species. In addition, we provide a phylogeny of Viburnum with Korean species, onto which some important morphological characters are mapped.
Materials and MethodsTaxon and gene samplingTen taxa distributed in Korea were included in this study (Table 1, Appendix 1). Viburnum erosum var. taquetii (H. Lév) Rehder was not included due to a lack of appropriate samples. Taxonomic identity of this taxon should be evaluated further, as a diagnostic character of V. erosum var. taquetii appears to be plastic. Four to eight accessions per taxon were analyzed, except for V. koreanum, whose distribution is limited at the high elevation of Mt. Seoraksan in Korea (Table 1). Viburnum clemensiae Kern was included as an outgroup based on previous studies (Winkworth and Donoghue, 2004; Clement and Donoghue, 2012; Clement et al., 2014).
Dried leaves of herbarium specimens or fresh leaves collected in the wild were used to isolate total DNA with the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Three cpDNA regions, in this case the rbcL, matK, and psbA-trnH regions and nuclear ribosomal ITS regions were amplified via polymerase chain reactions (PCR). Primers for the cpDNA regions from previous studies were used (Soltis et al., 1992; Sang et al., 1997; Cuénoud et al., 2002; Kress and Erickson, 2007; Kim et al., 2014). The ITS regions were amplified using the primers its6 and its9 (Youm et al., 2016). For each PCR reaction, 1 μL of total DNA was included in a 20 μL reaction mixture with Solg EF-Taq DNA polymerase (Solgent, Daejeon, Korea). Amplification of the target regions was conducted with a Veriti thermal cycler (Applied Biosystems, Foster City, CA, USA) under the following conditions: initial denaturation at 95°C for 2 min; 35 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; followed by a final extension at 72°C for 5 min. PCR products were examined on a 1% agarose gel in 1× TBE buffer, after which they were purified and sent to Solgent or Bionics (Seoul, Korea) for sequencing. Sequencing reaction used the PCR primers with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems).
For certain accessions of V. erosum, V. japonicum C. K. Spreng., and V. wrightii, direct sequencing results produced stuttering of the base readings and revealed the presence of multiple superimposed peaks. PCR products of those accessions were cloned using the T-Blunt PCR cloning kit (Solgent). Potentially transformed white colonies were incubated in Terrific Broth (Sambrook et al., 1989) at 37°C overnight, and plasmid DNA was isolated using the QIAprep Spin Miniprep kit (Qiagen). Two to four transformed clones per accession were sequenced with M13R and T7 primers. Sequences were determined using a 3730xl DNA analyzer (Applied Biosystems). Sequences were edited in Sequencher version 5.0 (Gene Codes Corporation, Ann Arbor, MI, USA), aligned manually.
Data analysisMultiple sequences were aligned using MUSCLE (Edgar, 2004) and were adjusted manually as needed. The three chloroplast regions were concatenated and analyzed simultaneously. The ITS data were analyzed separately and in combination with the cpDNA data. With regard to the accessions for which ITS sequences were determined by cloning, one sequence per accession was randomly selected and included in the combined data. Viburnum clemensiae was used as an outgroup. Phylogenetic analyses were conducted using the maximum parsimony (MP) criterion in PAUP* (Swofford, 2002) and Bayesian method (Huelsenbeck and Ronquist, 2001). All characters were treated as unordered and were weighted equally in the MP analyses. Gaps resulting from multiple alignments of indels were treated as missing. Heuristic searches were used with 100 replicates of random sequence additions with tree bisection-reconnection branch swapping, with all of the best trees saved at each step (MulTrees). Bootstrap analyses (Felsenstein, 1985) of 500 pseudoreplicates were conducted for cpDNA and the combined data with heuristic searches and a simple sequence addition in PAUP* to evaluate the support for each clade. “Fast bootstrap” with 50,000 pseudoreplicates was used for the ITS data.
Bayesian phylogenetic analyses were performed with the program MrBayes version 3.2.5 (Huelsenbeck and Ronquist, 2001). A Markov chain Monte Carlo algorithm was employed for 1,000,000 generations, sampling trees every 100 generations, with four independent chains running simultaneously. Priors were set based on the Akaike information criterion using Modeltest (Posada and Crandall, 1998). The first 2,000 trees (200,000 generations) were discarded as “burn-in,” and the remaining trees for which the log-likelihood values had reached a plateau were imported in PAUP* to calculate the posterior probability of each clade.
We examined key morphological characteristics using the materials collected in field and herbarium specimens deposited in KB (the herbarium of the National Institute of Biological Resources). Voucher specimens for the cross-section of stone to investigate presence of grooves of the stone surfaces and their numbers are provided in Appendix 2.
ResultsThe lengths of the rbcL, matK, and psbA-trnH regions were 565, 814, and 383–505, respectively. The final alignment of the cpDNA data includes 1,884 sites, among which 51 were parsimoniously informative (2.7%). The ITS data consist of 636 sites, of which 85 were parsimoniously informative (13.4%). The alignment of the combined data of the Korean Viburnum included 51 sequences with 2,517 sites, among which 134 were parsimoniously informative (5.3%). The summary of data and MP analysis were provided in Table 2. Best-fitting model for molecular evolution was selected as the TIMef model with equal rate for all three data sets.
Phylogenetic analysis of the cpDNA data resolved well-supported clades of species (Fig. 1). Four accessions of V. burejaeticum were supported as monophyletic, and those of V. carlesii Hemsl. formed a clade with V. burejaeticum. Monophyly of V. carlesii was not supported in the cpDNA data. Very low levels of variation among V. dilatatum Thunb., V. erosum, V. japonicum, and V. wrightii (hereafter the V. dilatatum complex) were found. The cpDNA data failed to differentiate the species in the V. dilatatum complex (Fig. 1). Most accessions of V. erosum and V. dilatatum, three of nine accessions of V. wrightii, and all accessions of V. japonicum showed the same sequences. Five accessions of V. wrightii and one of V. dilatatum were resolved as a sister to the remaining accessions of the species complex. DNA sequences from plants collected in Japan (V. japonicum Kyushu 043 and V. wrightii Kyushu 003) were identical to those plants in Korea. Viburnum koreanum was supported as a sister to V. opulus var. calvescens (Rehder) H. Hara. Accessions of V. odoratissimum var. awabuki (K. Koch) Zabel ex Rümpler and V. furcatum formed separate clades supporting the monophyly of each species (Fig. 1). Viburnum odoratissimum var. awabuki was a sister to the clade of the V. dilatatum complex and V. koreanum + V. opulus (Fig. 1). These species were resolved as a sister clade to the V. burejaeticum and V. carlesii clade. In our analysis of the cpDNA data, V. furcatum was supported as a sister to all of the Korean species (Fig. 1). One accession of V. furcatum collected in Hokkaido, Japan was differentiated from the Korean accessions.
The ITS data also showed similar patterns in the cpDNA data (Fig. 2). Noteworthy differences were that V. furcatum was a sister to V. odoratissimum var. awabuki, albeit low support values, and that the accessions of V. carlesii were strongly supported as monophyletic. All sequences of V. japonicum except for accession 7216 formed a cohesive unit, a sister to some of the accessions of V. erosum (Fig. 2). A phylogenetic analysis of the combined data was consistent with the ITS tree (Fig. 3).
The species resolution power, by which each species was differentiated in the phylogenetic tree, was calculated and found to be 60% in separate analyses of the cpDNA and ITS data and in the combined analysis. Viburnum burejaeticum, V. carlesii, V. koreanum, V. opulus var. calvescens, V. odoratissimum var. awabuki, and V. furcatum were distinguished by any of the sequence data. However, the DNA barcode sequences did not differentiate the four species of the V. dilatatum complex.
DiscussionThe results of our phylogenetic analysis of three chloroplast regions (Fig. 1) are consistent with the findings of previous studies (Donoghue et al., 2004; Winkworth and Donoghue 2004, 2005; Clement and Donoghue 2011, 2012; Clement et al., 2014). Classification of Viburnum using phylogenetic nomenclature (Clement et al., 2014) was based on a phylogenetic analysis of the chloroplast genome, which recognized four major clades, V. clemensiae, Valvatotinus, Pseudotinus, and Pluriviburnum. Our analysis of the cpDNA data supports these clades (Fig. 1). Viburnum burejaeticum and V. carlesii classified in sect. Viburnum (Hara, 1983) are recognized in Euviburnum in the phylogenetic nomenclature (Clement et al., 2014). Euviburnum along with Lentago and Punctata belong to a more inclusive clade of Valvatotinus. Viburnum furcatum is a member of Pseudotinus. Pluriviburnum is a large clade containing most of the diversity of Viburnum (Clement et al., 2014). The Korean taxa, specifically the V. dilatatum complex, V. koreanum, and V. opulus var. calvescens, belong to Pluriviburnum. The phylogenetic relationships in our analysis of the cpDNA data suggested that Valvatotinus is more closely related to Pluriviburnum than it is to Pseudotinus with a low bootstrap support value (Fig. 1). Valvatotinus was suggested as a sister clade of Pseudotinus in previous studies (Clement et al., 2014). However, support values for a close relationship between Valvatotinus and Pseudotinus were low (Clement et al., 2014).
A phylogenetic analysis of the ITS regions placed Viburnum odoratissimum var. awabuki as a sister to V. furcatum with relatively high bootstrap support (Fig. 2). The results of a combined analysis of the cpDNA and ITS regions are consistent with the ITS data (Fig. 3), suggesting that phylogenetic signals from the ITS regions are stronger than cpDNA. Previous analysis of the ITS region (Donoghue et al., 2004) and nuclear GBSSI (Winkworth and Donoghue, 2004) also showed a close relationship between Pseudotinus and Solenotinus, to which Viburnum odoratissimum belongs.
Viburnum furcatum is characterized by having a pair of caducous bud scales resulting in naked buds at a later stage (from late autumn to spring), umbellate inflorescences with marginal sterile flowers, and one deep ventral groove in the stones (Fig. 4). Our DNA sequence data indicate that the species can be clearly discriminated from other species (Figs. 1, 2). It is distributed from South Sakhalin, Japan and to Korea and then to Taiwan (Hara, 1983; Kim, 2007). Although it is common in Japan, its distribution in Korea is restricted, occurring at rather high elevations on Mt. Hallasan and on Ulleungdo Island. A sporadic occurrence of the species at Mt. Jabyeongsan in Gangwon province in Korea was reported (Son et al., 2008). There is little molecular variation within the species (Figs. 1, 2). The ITS regions were identical across the Korean accessions examined in this study. ITS sequence from the plant from Hokkaido (accession number 7464) had unique insertion. The Japanese accession was distinguished from the plants on the Jejudo Island in the cpDNA data (Fig. 1), suggesting that there may have been haplotype diversity with the species. We did not sample plants from Ulleungdo Island or Mt. Jabyeongsan in this study. The cpDNA regions and perhaps more variable regions will be useful to infer the origins of the populations.
Viburnum odoratissimum var. awabuki is found on Jejudo Island in Korea and in Japan, Taiwan, and the Philippines. This species differs from others by having a basic chromosome number of x = 8, which is rare in Viburnum (Zhang et al., 2016). Most other species of Viburnum have a basic chromosome number of x = 9 (Egolf, 1962). The species is easily recognized by having subentire leaves and paniculate inflorescences borne on evergreen trees (Fig. 4). Our DNA barcodes clearly differentiate the species (Figs. 1, 2).
Viburnum burejaeticum is distributed at high latitudes in Korea, the Russian Far East, and northeastern China. We included samples from Gangwon-do Province in Korea and Jilin in China (Table 1), and there was no intra-specific variation in both the cpDNA and ITS regions (Figs. 1, 2). This species is morphologically distinguished by naked buds and exerted stamens from rotate corolla and is often confused with V. dilatatum when flowers are unavailable. However, V. burejaeticum does not have an extrafloral nectary, whereas V. dilatatum has an extrafloral nectary on the abaxial surface of the leaves (Fig. 4). Viburnum carelsii was considered to be closely related to V. burejaeticum by having naked buds and stones with two dorsal and three ventral shallow grooves (Hara, 1983). The two species represent the Euviburnum clade (Clement et al., 2014). Our molecular data show that the two species can be discriminated (Figs. 1, 2). Viburnum carelsii can be easily distinguished from V. carlesii by having hypocrateriform corollas in which stamens are included.
Viburnum koreanum and V. opulus are members of the sect. Opulus (Hara, 1983). Our data with multiple accessions from different populations of V. opulus in Korea resolved the two species as distinct (Figs. 1, 2). The presence of an extrafloral nectary on the upper part of the petiole is a synapomorphy for Opulus (Clement et al., 2014). All materials examined here showed these characteristics. Viburnum koreanum, distributed in northern and central Korea and in Manchuria and Hokkaido is rare throughout the range, while V. opulus is common. All specimens labelled as V. koreanum deposited in KB were incorrectly identified; they were V. opulus. Viburnum koreanum can be distinguished from V. opulus by having sharply serrate leaves and smaller inflorescences lacking marginal sterile flowers (Hara, 1983). Hara (1983) reported that plants from Hokkaido lack a petiolar extrafloral nectary. This can be explained as a secondary loss of the trait within a species.
The V. dilatatum complex, which consists of four closely related species, is characterized by free bud scales and serrate leaves with pinnate veins and extrafloral nectaries at proximal regions on the abaxial surface. The complex formed a highly supported clade in both cpDNA and ITS trees (Figs. 1, 2). Relationships among the species within the complex are unresolved, however. The DNA barcodes examined in this study could not differentiate the species. This suggests that it is difficult to infer a species boundary from the sequence data in the V. dilatatum complex. A previous cpDNA analysis with 113 species of Viburnum found that V. dilatatum is a sister to a clade that includes V. erosum, V. wrightii, and V. japonicum with low support values (Clement et al., 2014). Our cpDNA data show that some accessions of V. wrightii are differentiated from the remaining three species (Fig. 1). However, other accessions of V. wrightii possess a haplotype identical to those of the other species. Likewise, four of six accessions of V. erosum formed with a distinct clade in the ITS data but the remaining two accessions were nested within the V. dilatatum clade (Fig. 2). All accessions of V. dilatatum except for J9050 formed a cohesive unit particularly in the ITS data (Fig. 2). The outlier accession J9050 was placed outside of the V. dilatatum clade and had the same cpDNA haplotype as V. wrightii. Incomplete lineage sorting may have played a role in the patterns observed in the DNA barcodes in the V. dilatatum complex.
The species in the complex can be differentiated by morphological characteristics. Viburnum japonicum is easily distinguished from other Korean species in the V. dilatatum complex by having glabrous evergreen leaves. This species is distributed in warm areas of southern Japan from Honshu, Kyushu to the Ryukyu Islands. The distribution range has recently been expanded to include Gageodo Island in southwestern Korea (Hong and Im, 2003). Viburnum erosum is distributed in southern Korea and offshore islands in western Korea; western Japan; and southern China. It can be distinguished by having stipules. None of the other species of Viburnum in Korea have stipules. Pubescence on branchlets has been used to distinguish the four species (Kim, 2007). Branchlets of V. japonicum are glabrous, and those of V. wrightii are usually glabrous or occasionally pubescent with long simple hairs. Branchlets of V. erosum have stellate hairs, whereas those of V. dilatatum are densely pubescent with stellate and simple hairs.
Our analyses of DNA barcode regions from multiple accessions of the species of Viburnum in Korea confirm that six out of ten species in Korea can be discriminated at the species level. The V. dilatatum complex can be separated from the remaining species according to molecular data, but the resolution power to differentiate a species within the complex is absent in our data. This study suggests that regional DNA barcodes are useful for molecular species identification for Viburnum when flowering or fruiting materials are not available.
ACKNOWLEDGMENTSWe thank Narae Yun, Seung Hyun Hwang, and Ji-Hyun Park for their help with the fieldwork, and we thank Dong-Hyuk Lee and Hwa-Jung Suh for obtaining the plant samples. We are grateful to two anonymous reviewers for their critical reviews of the manuscript. This work was supported by a research grant, NIBR-201614101, from the National Institute of Biological Resources, Korea.
Fig. 1.One of four maximum parsimony tress from a phylogenetic analysis of the concatenated data of rbcL, matK, and psbA-trnH regions from Viburnum in Korea. Bootstrap values greater than 50% are indicated above the corresponding branch and posterior probability in Bayesian analysis was below. The accession number follows the species name to distinguish each individual. Viburnum clemensiae was used as an outgroup. 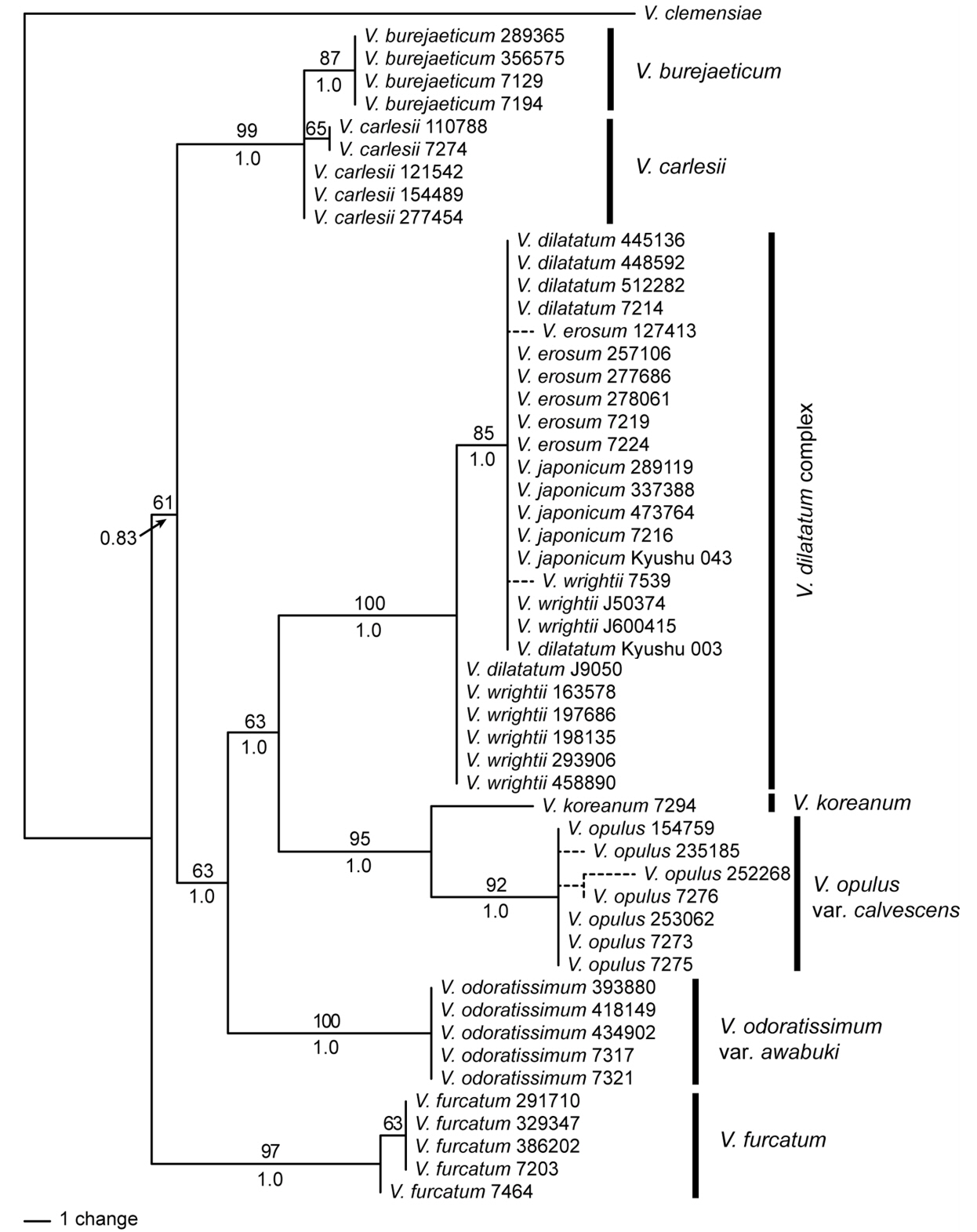
Fig. 2.One of 2,075 maximum parsimony trees from a phylogenetic analysis of the internal transcribed spacer regions from Viburnum in Korea. Dotted lines represent branches collapsed in the strict consensus tree. Bootstrap values greater than 50% are indicated above the corresponding branch and posterior probability in Bayesian analysis was below. Accession numbers are indicated following the species name to distinguish each individual. In some accessions of V. erosum and V. japonicum, the sequences were determined by cloning. The clone number follows the accession number. Viburnum clemensiae was used as an outgroup. 
Fig. 3.Strict consensus tree of 2,231 maximum parsimony trees from a phylogenetic analysis of the combined cpDNA and internal transcribed spacer (ITS) data from Viburnum in Korea. Bootstrap values greater than 50% are indicated above the corresponding branch and posterior probability in Bayesian analysis was below. The accession number follows the species name to distinguish each individual. For cloned ITS sequences, one randomly selected clone per accession was included in the combined data. Viburnum clemensiae was used as an outgroup. 
Fig. 4.Summary phylogeny of Viburnum in Korea with a comparison of key morphological characteristics. Character A, presence or absence of bud scales; B, habit of plants; C, leaf margin; D, distribution of extrafloral nectary; E, inflorescence type; F, fruit color in maturity, G, number of grooves on the dorsal surface and ventral surface of the stone. An exemplar cross-section of the stone is provided following the character G to show the dorsal and ventral groove, if present. An asterisk is used to indicate that plants in Hokkaido lack a petiolar extrafloral nectary. 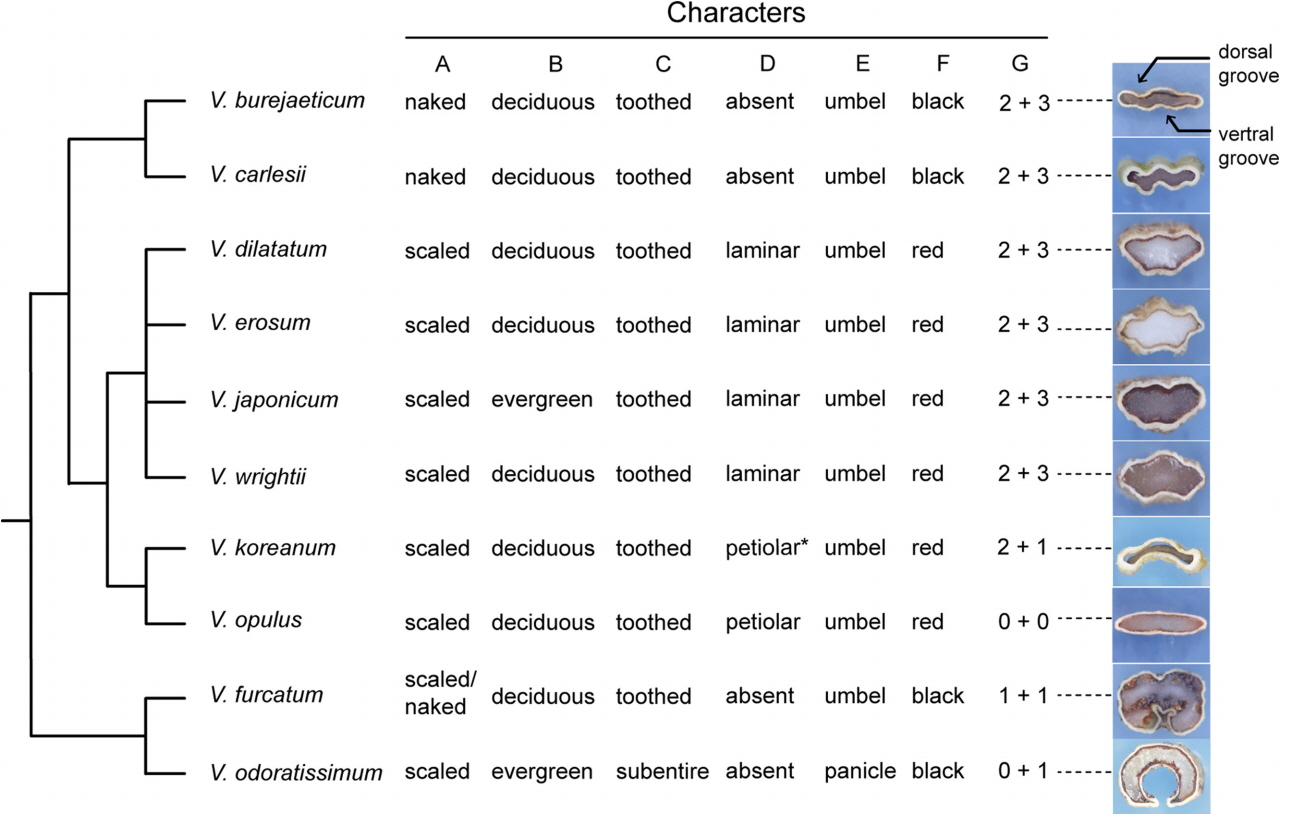
Table 1.List of species included in this study with higher classification categories and information of the sources of materials. Sectional classification of Viburnum follows Hara (1983) and clade name Clement et al. (2014). CB, Chungcheongbuk-do; GB, Gyeongsangbuk-do; GG, Gyeonggi-do; GW, Gangwon-do; JB, Jeollabuk-do; JJ, Jeju-do; JN, Jeollanam-do. Table 2.Summary statistics of cpDNA, ITS, and combined data as used in our phylogenetic study of Viburnum. Literature CitedAhn, DK. 1998. Illustrated Book of Korean Medicinal Herbs. Kyohaksa, Seoul. 854 pp.
Cantino, PD. Doyle, JA. Graham, SW. Judd, WS. Olmstead, RG. Soltis, DE. Soltis, PS and Donoghue, MJ. 2007. Towards a phylogenetic nomenclature of Tracheophyta.. Taxon 56: 822-846.
CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the United States of America 106: 12794-12797.
Clement, WL. Arakaki, M. Sweeney, PW. Edwards, EJ and Donoghue, MJ. 2014. A chloroplast tree for Viburnum (Adoxaceae) and its implications for phylogenetic classification and character evolution.. American Journal of Botany 101: 1029-1049.
Clement, WL and Donoghue, MJ. 2011. Dissolution of Viburnum section Megalotinus (Adoxaceae) of Southeast Asia and its implications for morphological evolution and biogeography.. International Journal of Plant Sciences 172: 559-573.
Clement, WL and Donoghue, MJ. 2012. Barcoding success as a function of phylogenetic relatedness in Viburnum, a clade of woody angiosperms. BMC Evolutionary Biology 12: 73.
Cuénoud, P. Savolainen, V. Chatrou, LW. Powell, M. Grayer, RJ and Chase, MW. 2002. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences.. American Journal of Botany 89: 132-144.
Donoghue, MJ. 1983. The phylogenetic relationships of Viburnum
. Advances in Cladistics 2: 143-166.
Donoghue, MJ. Baldwin, BG. Li, J and Winkworth, RC. 2004.
Viburnum phylogeny based on chloroplast trnK intron and nuclear ribosomal ITS DNA sequences.. Systematic Botany 29: 188-198.
Donoghue, MJ. Bell, CD and Winkworth, RC. 2003. The evolution of reproductive characters in Dipsacales.. International Journal of Plant and Science 164: S453-S464.
Edgar, RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792-1797.
Egolf, DR. 1962. A cytological study of the genus Viburnum
. Journal of the Arnold Arboretum 43: 132-172.
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783-791.
Hara, H. 1983. A Revision of Caprifoliaceae of Japan with Reference to Allied Plants in Other Districts and the Adoxaceae. Academia Scientific Books, Tokyo. 336 pp.
Hebert, PDN. Cywinska, A. Ball, SL and deWaard, JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences. 270: 313-321.
Hong, H-H and Im, H-T. 2003.
Viburnum japonicum (Caprifoliaceae): an unrecorded species in Korea. Korean Journal of Plant Taxonomy 33: 271-277.
Huelsenbeck, JP and Ronquist, F. 2001. MrBayes: Bayesian inference of phylogenetic tress. Bioinformatics 17: 754-755.
Kim, HM. Oh, S-H. Bhandari, GS. Kim, C-S and Park, C-W. 2014. DNA barcoding of Orchidaceae in Korea. Molecular Ecology Resources 14: 499-507.
Kim, TJ. 2007. Viburnaceae Raf. The Genera of Vascular Plants of Korea. Park, C-W (ed.), Academy Publishing Co, Seoul. 944-946.
Kress, WJ and Erickson, DL. 2007. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region.. PLoS ONE 2: e508.
Kress, WJ. Wurdack, KJ. Zimmer, EA. Weigt, LA and Janzen, DH. 2005. Use of DNA barcodes to identify flowering plants.. Proceedings of the National Academy of Sciences of the United States of America 102: 8369-8374.
Kwon, MK. Lee, HE. Park, JY and Hahn, YS. 2003. Antimicrobial activities of berry extract of domestic plants on 4 kinds of pathogenic microorganism.. Journal of the East Asian Society of Dietary Life 13: 433-438.
Lahaye, R. van der Bank, M. Bogarin, D. Warner, J. Pupulin, F. Gigot, G. Maurin, O. Duthoit, S. Barraclough, TG and Savolainen, V. 2008. DNA barcoding the floras of biodiversity hotspots.. Proceedings of the National Academy of Sciences of the United States of America 105: 2923-2928.
Mabberley, DJ. 1997. The Plant-Book. 2nd ed. Cambridge University Press, Cambridge. 680 pp.
Posada, D and Crandall, KA. 1998. MODELTEST: testing the model of DNA substitution.. Bioinformatics 14: 817-818.
Rehder, A. 1908. The Viburnums of eastern Asia. Trees and Shrubs. Vol. II, Part II. Sargent, CS (ed.), Houghton Mifflin, Boston, MA. 105-116.
Sambrook, J. Fritzsch, EF and Maniatis, T. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press, New York. 2100 pp.
Sang, T. Crawford, D and Stuessy, T. 1997. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany 84: 1120-1136.
Soltis, PS. Soltis, DE and Smiley, CJ. 1992. An rbcL sequence from a Miocene Taxodium (bald cypress).. Proceedings of the National Academy of Sciences of the United States of America 89: 449-451.
Son, H-D. Kim, D-H and Im, H-T. 2008. A short record for the distribution of 4 rare plants. Korean Journal of Plant Taxonomy 38: 565-571.
Swofford, DL. 2002. PAUP* Phylogenetic analysis using parsimony (* and other methods), version 4.0. Sinauer Associates, Sunderland, MA.
Winkworth, RC and Donoghue, MJ. 2004.
Viburnum phylogeny: evidence from the duplicated nuclear gene GBSSI
. Molecular Phylogenetics and Evolution 33: 109-126.
Winkworth, RC and Donoghue, MJ. 2005.
Viburnum phylogeny based on combined molecular data: implications for taxonomy and biogeography.. American Journal of Botany 92: 653-666.
Xiang, X-G. Hu, H. Wang, W and Jin, X-H. 2011. DNA barcoding of the recently evolved genus Holcoglossum (Orchidaceae: Aeridinae): a test of DNA barcode candidates. Molecular Ecology Resources 11: 1012-1021.
APPENDICESAppendix 1.List of samples included in phylogenetic study
Appendix 2.List of samples used in anatomical examination of the stones
|
|
|||||||||||||||||||||||||||||||||||