Abstract
Paraphlomis koreana (Lamiaceae) was newly named and added to Korean flora in 2014. Paraphlomis belongs to the tribe Paraphlomideae, along with Ajugoides and Matsumurella. However, a recent study has suggested that P. koreana is morphologically similar to Matsumurella chinensis, making them difficult to distinguish from each other. Therefore, we aimed to examine the phylogenetic placement of P. koreana within the tribe and compare its genetic relationship with M. chinensis. We sequenced an additional complete plastid genome for an individual of P. koreana and generated sequences of nuclear ribosomal (nr) DNA regions of internal and external transcribed spacers (ITS and ETS) for two individuals of P. koreana. Maximum likelihood analyses based on two nrDNA regions (ITS and ETS) and four plastid DNA markers (rpl16 intron, rpl32-trnL, rps16 intron, and trnL-F) covering 13 Paraphlomis species and M. chinensis were conducted. Phylogenetic analyses concordantly supported that P. koreana forms a monophyletic group with M. chinensis. Moreover, our study revealed that P. koreana includes nrDNA sequences of M. chinensis as minor intraindividual variants, suggesting that the genetic divergence between the two taxa is incomplete and may represent intraspecific variation rather than distinct species. In conclusion, our findings suggest that the independent species status of P. koreana within Paraphlomis should be reconsidered.
INTRODUCTIONParaphlomideae Bendiksby is a tribe within Lamiaceae that consists of three genera: Ajugoides Makino, Matsumurella Makino, and Paraphlomis Prain (Bendiksby et al., 2011). Recently, a plastid genome (plastome) phylogenomics-based classification of Lamiaceae recognized this tribe as a distinct lineage within the subfamily Lamioideae with a combination of some morphological traits (Zhao et al., 2021). Ajugoides is a monotypic genus endemic to Japan (Makino, 1915) while Matsumurella contains five species distributed in China, Japan, and Taiwan (Li and Hedge, 1994; Bendiksby et al., 2011; Tagane and Nakanishi, 2021). Paraphlomis, on the other hand, has around 36 species, eight of which were newly described species in the last decade (Ko et al., 2014; Zhang et al., 2020; Chen et al., 2021, 2022a, 2022b; Yuan et al., 2022; Zhao et al., 2022; Guo et al., 2023).
Paraphlomis koreana S. C. Ko et G. Y. Chung, which was discovered on Bogil-do Island, is the only representative of the tribe and genus in Korea (Ko et al., 2014; Chung et al., 2023). Ko et al. (2014) proposed that P. koreana can be distinguished from other congeners based on some key features (height, leaf, bracteole, flower, and calyx) and suggested that Paraphlomis albida Hand.-Mazz. is its closest relative. However, Tagane and Nakanishi (2021) pointed out that P. koreana is quite similar to M. chinensis in gross morphology, which raises questions about the taxonomic status of the former as an independent species.
A recent phylogenomic analysis of P. koreana (Park et al., 2022) only included one additional Paraphlomis species, Paraphlomis javanica (Blume) Prain. However, a preliminary phylogenetic study of Paraphlomis (Chen et al., 2021) utilized two nuclear ribosomal (nr) DNA regions of internal and external transcribed spacers (ITS and ETS) and four plastid DNA markers (rpl16 intron, rpl32-trnL, rps16 intron, and trnL-F) and included 15 Paraphlomis species (excluding P. koreana) and M. chinensis. In this study, we aimed to determine the phylogenetic position of P. koreana within the Paraphlomideae tribe, specifically its relationship with Paraphlomis and Matsumurella species.
MATERIALS AND METHODSPlant material sampling and next-generation sequencingFresh leaves of an individual of P. koreana were collected from natural habitats in Bogil-myeon, Wando-gun, Jeollanam-do, South Korea (Fig. 1). The voucher specimen (J.-H. Kim KIMJH22027) was deposited at the herbarium of the National Institute of Biological Resources (KB) with sample number NIBRVP0000829804. Total genomic DNA was extracted from silica-gel-dried leaves using the DNeasy Plant Mini Kit (Qiagen, Seoul, Korea) according to the manufacturer’s protocol. The genomic library was constructed using TruSeq Nano DNA Kit and sequenced in 151-bp paired-end reads on Illumina Hiseq platform (Macrogen, Seoul, Korea).
In this study, raw next-generation sequencing (NGS) data and its derivatives of two P. koreana individuals from this study and a previous study (Park et al., 2022) were discriminated by adding suffixes after the scientific name (P. koreana_1 = D.C. Son Bogildo-200716-001; P. koreana_2 = J.-H. Kim KIMJH22027).
Plastid genome assembly and comparisonA plastome of P. koreana_2 was assembled using GetOrganelle v1.7.4.1 (Jin et al., 2020). The plastome assemblage was confirmed by paired-end read mapping using the low sensitivity option in Geneious Prime 2022.1.1 (https://www.geneious.com/). The completed plastome was annotated using references of P. koreana_1 (NC_058605.1) and P. javanica (MT473773.1) in Geneious Prime. Complete plastomes of two P. koreana individuals were compared by sequence alignment using MAFFT v.7.450 (Katoh and Standley, 2013), and the alignment algorithm was automatically selected.
Assembly of nuclear ribosomal DNA regions and detection of intraindividual sequence polymorphismThe NGS data from P. koreana_1 generated by Park et al. (2022) was downloaded from the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) of the National Center for Biotechnology Information (NCBI). To assemble nrDNA regions of two individuals of P. koreana, each NGS data file was mapped to reference ITS (MW602147) and ETS (MW602117) sequences of M. chinensis (Chen et al., 2021) in Geneious Prime with medium-low sensitivity. If one read of a pair of unmapped reads was mapped, unmapped reads were also saved. Saved reads were assembled in Geneious Prime with medium-low sensitivity. Intraindividual sequence polymorphism of 0.1 minimum variant frequency was checked using the “Find Variations/SNPs” function in Geneious Prime.
Phylogenetic analysesSequences of two nrDNA (ITS and ETS) and four plastid markers (rpl16 intron, rpl32-trnL, rps16 intron, and trnL-F) generated by Chen et al. (2021) were acquired from GenBank. From nrDNA assemblies and plastomes of P. koreana, corresponding sequences to the six DNA regions were extracted. For two nrDNA regions of P. koreana, majority consensus sequences were used. Each DNA region was aligned using MAFFT as described above. Three datasets were prepared for phylogenetic analyses: (1) nrDNA, (2) plastid DNA, and (3) combined DNA (all nr and plastid DNA regions) (Appendix 1). Maximum likelihood analyses were conducted using IQ-TREE 2.2.0 (Minh et al., 2020) with 10,000 bootstrap replications. The best partition scheme was determined, and the Bayesian Information Criterion was used to select a best-fit nucleotide substitution model.
Distribution survey of Paraphlomis koreana and Matsumurella chinensisGeographical distribution data for the target taxa, P. koreana and M. chinensis, were obtained from literature sources and the Global Biodiversity Information Facility (GBIF; https://www.gbif.org/; accessed July 28, 2022). The distribution of P. koreana was confirmed through published literature (Ko et al., 2014; Park et al., 2022), while that of M. chinensis was identified from the GBIF occurrence database. Geo-referenced record data were downloaded from GBIF, which included data on Galeobdolon chinense (Benth.) C. Y. Wu and Lamium chinense Benth., which are homotypic synonyms of M. chinensis. To minimize misidentification, we excluded data solely based on human observations. We also removed occurrences from locations outside the known distributional range of the species, as listed in Flora of China (Li and Hedge, 1994). Finally, a distribution map was generated using QGIS 3.22 software based on carefully selected data obtained from occurrences and preserved specimens (such as herbarium vouchers).
RESULTSPlastid genomes of two Paraphlomis koreana individuals are identicalThe plastid genome of P. koreana_2 (accession number OQ842967) was sequenced at a read coverage of 3,748-fold and its length was determined to be 152,128 bp. The sequence of this newly assembled plastome from the same population was identical to a previously sequenced plastome (P. koreana_1; NC_058605).
Nuclear ribosomal and plastid data support the close phylogenetic relationship between Paraphlomis koreana and Matsumurella chinensisThree phylogenetic trees were constructed using (1) nrDNA (Fig. 2A), (2) plastid DNA (Fig. 3A), and (3) combined DNA data (Fig. 4). The results indicated that both P. koreana and M. chinensis formed monophyletic groups. The monophyly of the two taxa was maximally supported with a bootstrap support value (BS) of 100 in all analysis. However, the sister group of this monophyletic group of the two taxa differed in the three analyses. Paraphlomis seticalyx C. Y. Wu was identified as the sister species in the trees based on nrDNA (BS = 71) and combined DNA (BS = 57) data. Due to the phylogenetic position of Matsumurella, the Paraphlomis was not monophyletic.
Assessment of intraindividual variation in nuclear ribosomal DNA shows incomplete sequence divergence between Paraphlomis koreana and Matsumurella chinensisIn the alignment of nrDNA sequences (1,072 bp in total, aligned length) from four individuals of P. koreana and M. chinensis, nine polymorphic sites were identified. One individual of M. chinensis_1 displayed mixed sequences at three sites, with bases of W (A or T) or Y (C or T). Two individuals of P. koreana were found to have six intraindividual polymorphic sites of nrDNA, as shown in Fig. 2B. The phylogenetic analysis, based on the major variant of nrDNA, revealed a sequence divergence between P. koreana and M. chinensis (see Fig. 2A). However, when considering the minor variants, no species-specific mutations were found. For instance, the T nucleotide at aligned position 384 of M. chinensis was also present as minor variants in the two individuals of P. koreana.
Interindividual plastid sequence variation in the rpl16 intron, rpl32-trnL, rps16 intron, trnL-trnF regions among four individuals of Paraphlomis koreana and Matsumurella chinensisWe examined the variation of the combined plastid sequence of the rpl16 intron, rpl32-trnL, rps16 intron, and trnL-trnF regions (3,252 bp in total, aligned length) (Fig. 3B). While the rpl16 intron sequences were identical among the four individuals of P. koreana and M. chinensis, other regions showed some indels and substitutions. In the rpl32-trnL region, no sequence substitution was observed, but two deletions of a nucleotide and 146 bp were found in M. chinensis. The 146 bp deletion in the rpl32-trnL region was specific to two individuals of M. chinensis among all examined taxa. In total, five variable sites of substitutions were found in the rps16 (one) and trnL-F (four) regions. One substitution was specific to an individual of M. chinensis, while the other substitution sites showed species-specific variations.
Distribution of Paraphlomis koreana and Matsumurella chinensisThe distribution of M. chinensis and closely related species was surveyed and is depicted in Fig. 5. Matsumurella chinensis was found to be distributed within the range of approximately 24.5 to 33.0°N and 109.0 to 129.5°E, with occurrences mainly concentrated in Southeastern China and Northern Taiwan. Notably, a disjunct population was observed on Goto Island, Japan, which is approximately 700 km away from the nearest occurrence in Zhejiang, China. Although the location of P. koreana (Bogil-do Island, Korea) falls within the longitudinal distribution range of M. chinensis, it is slightly outside the latitudinal range. Nonetheless, Bogil-do Island is relatively closer (approximately 650 km) to the occurrence of M. chinensis in Zhejiang, China, than Goto Island, Japan. The record of an undetermined lineage of Matsumurella (Matsumurella sp.) from Tsushima Island, Japan, was slightly further north in latitude than P. koreana.
DISCUSSIONThe generic position of Paraphlomis koreanaIn this study, we examined the phylogenetic position of P. koreana for the first time. Our phylogenetic analyses supported the monophyly of P. koreana and its close relationship to M. chinensis (Figs. 2–4) which is consistent with the view of Tagane and Nakanishi (2021) that P. koreana is morphologically similar to M. chinensis. Bendiksby et al. (2011) suggested that Matsumurella species can be distinguished from Paraphlomis by the length of their calyx lobes, which are over half as long as the tube. In this regard, the calyx lobes of P. koreana (Fig. 1E) resemble those of Matsumurella. However, it should be noted that the calyx lobes of Paraphlomis species are highly variable, and some species (P. gracilis and P. reflexa) exhibit a calyx lobes/tube ratio similar to that of Matsumurella (Chen et al., 2021). Furthermore, as demonstrated in our study and previous molecular phylogenetic studies (Zhang et al., 2020; Chen et al., 2021, 2022a, 2022b; Yuan et al., 2022; Zhao et al., 2022; Guo et al., 2023), Paraphlomis is not a monophyletic group since Matsumurella is nested within it. It is also known that a monotypic genus Ajugoides is morphologically less distinct from Matsumurella (Bendiksby et al., 2011; Chen et al., 2021). Therefore, while P. koreana exhibits a strong phylogenetic affinity with Matsumurella, it may not require a generic transfer since future taxonomic study for tribe Paraphlomideae could merge all three genera. More importantly, it should be re-confirmed that P. koreana is an independent species from currently recognized species in the tribe.
Interspecific relationship between Paraphlomis koreana and Matsumurella chinensis
Matsumurella chinensis exhibits a highly variable morphology, particularly in its leaves and corolla, and infraspecific taxa have been identified previously (Xiang et al., 2016). Although the species is the most similar to Matsumurella tuberifera (Makino) Makino, it can be distinguished by having ovate-oblong to broadly lanceolate leaves (Li and Hedge, 1994). It is difficult to distinguish between P. koreana and M. chinensis based on this diagnostic characteristic. However, our phylogenetic analyses recognized P. koreana as an independent lineage to M. chinensis with some sequence divergence (Figs. 2–4) even though other Matsumurella species were not sampled. In this regard, interspecific, interindividual, and intraindividual-level genetic variation among four individuals of P. koreana and M. chinensis are worthy of further discussion (Figs. 2, 3).
The major variants of nrDNA exhibited lineage divergence between the two taxa (Fig. 2A). However, P. koreana individuals included sequence types of M. chinensis as their minor variants (Fig. 2B). As the nrDNA exist as multi-loci tandemly repetitive units, ancestral remnant sequence could coexist with derived sequence generated from recent de novo mutations (Cardoni et al., 2022). Moreover, considering the distance between the sampling location of P. koreana (Bogil-do Island, Korea) (see Fig. 5) and M. chinensis (Jiangxi and Guangxi provinces, China) (Chen et al., 2021), the shared nrDNA sequence variation (Fig. 2B) may represent recent intraspecific-level divergence due to their geographic distance.
Plastid DNA usually maternally inherited in angiosperms (Corriveau and Coleman, 1988), and its gene flow is often restricted by movement of seeds (McCauley, 1995). Thus, plastid DNA variation often represent geography rather than taxonomy (Petit and Excoffier, 2009). Similarly, our analysis of plastid DNA (Fig. 3B) showed clear sequence divergence between P. koreana and M. chinensis, in contrast to the nrDNA results (Fig. 2B). This level of divergence is reminiscent of phylogeographic structuring of plant species around East China, the southern tip of Korea, and southern Japan (Qiu et al., 2011; Chen et al., 2012; Lee et al., 2013; Jin et al., 2016; Cao et al., 2018). Locality of P. koreana (Bogil-do Island, Korea) is slightly outside the latitudinal range of M. chinensis but is closer to occurrence from mainland China than Goto Island, Japan (see Fig. 5). When considering the available evidence from morphology, sequence divergence, and distribution, it is plausible that P. koreana and M. chinensis may be conspecific. However, phylogenetic analyses with enough sampling that represents multiple Matsumurella species and intraspecific diversity of M. chinensis are necessary to draw conclusion.
ACKNOWLEDGMENTSWe thank Mr. Kang-Hyup Lee at Korea National Arboretum for providing us the photo of Nutlet. This work was supported by a grant from the National Institute of Biological Resources (NIBR202203102).
Fig. 1.
Paraphlomis koreana S. C. Ko et G. Y. Chung. A. Habitat. B. Habit. C. Root with tuber. D. Stem. E. Corolla with calyx. F. Open corolla with stamens and pistil. G. Nutlet. 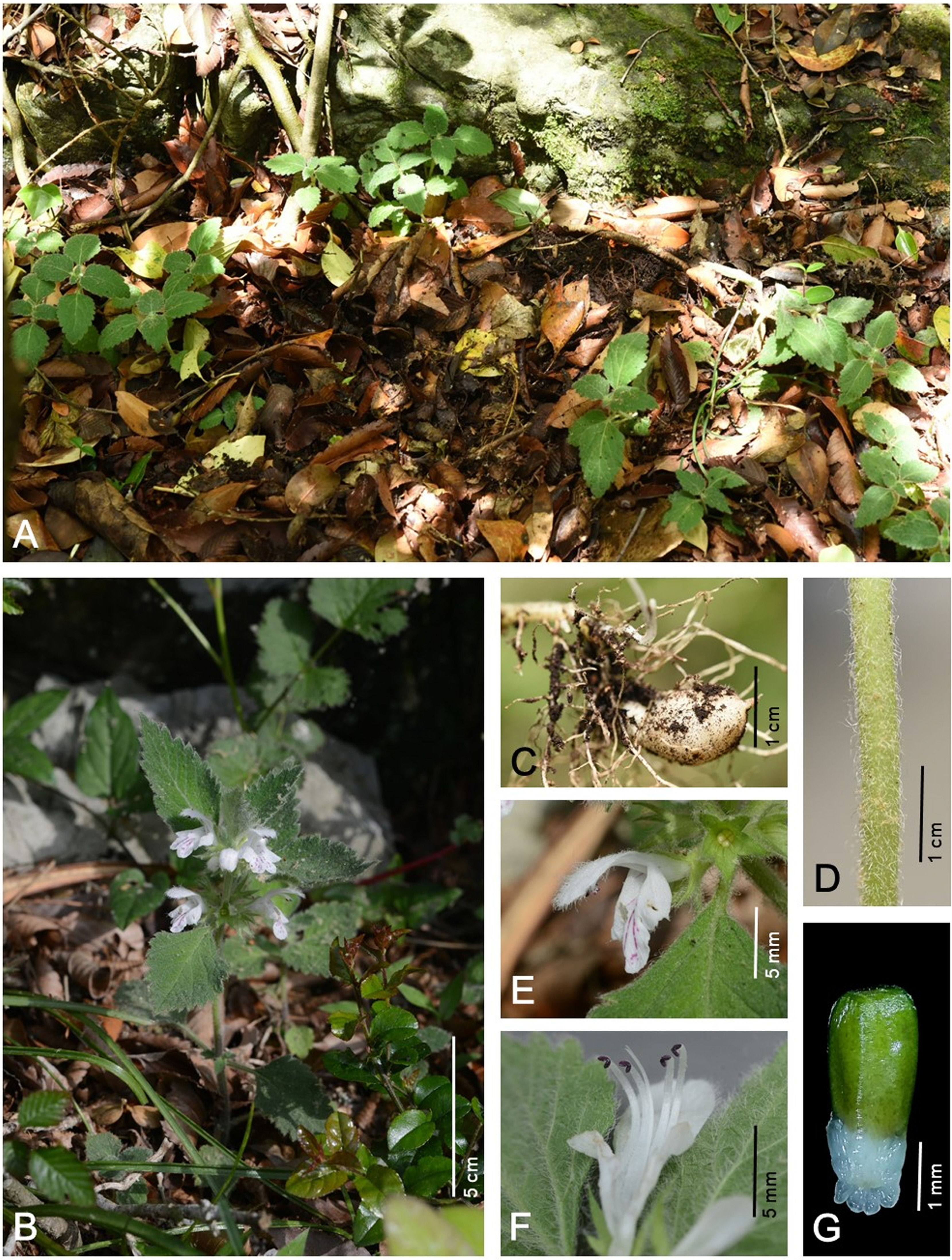
Fig. 2.Phylogenetic position of Paraphlomis koreana (A) and its sequence divergence from Matsumurella chinensis (B) inferred from nrDNA (ITS and ETS). Scale indicates the number of nucleotide substitutions per site. Bootstrap support values for each node are presented. The sequence alignment for P. koreana shows the proportion of alternative sequences in each locus, displayed within brackets. nrDNA, nuclear ribosomal DNA; ITS, internal transcribed spacer; ETS, external transcribed spacer. 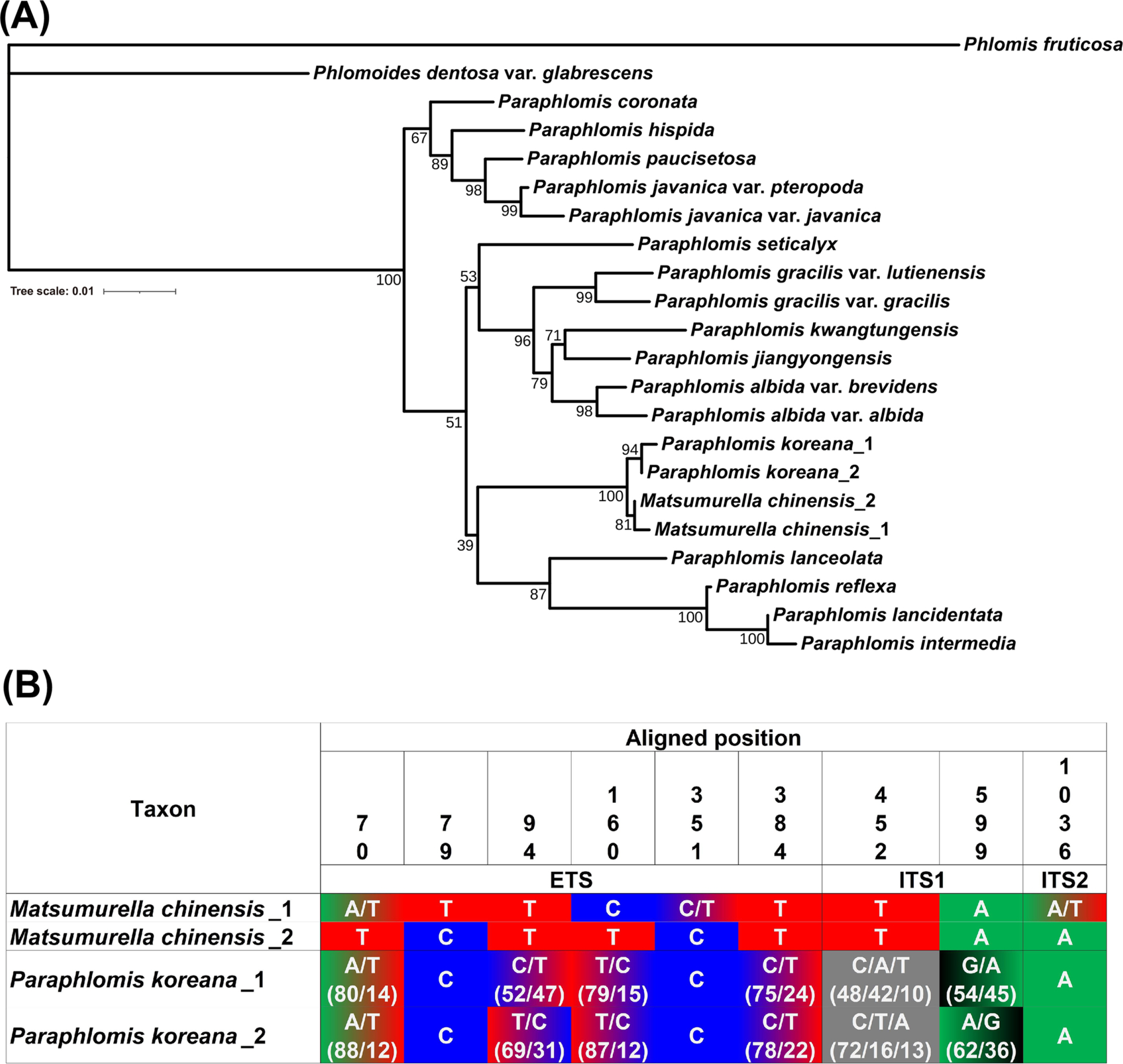
Fig. 3.Phylogenetic position of Paraphlomis koreana (A) and its sequence divergence from Matsumurella chinensis (B) inferred from plastid DNA regions (rpl16 intron, rpl32-trnL, rps16 intron, and trnL-F). Scale indicates the number of nucleotide substitutions per site. Bootstrap support values for each node are presented. 
LITERATURE CITEDBendiksby, M. Thorbek, L. Scheen, A.-C. Lindqvist, C and Ryding, O. 2011. An updated phylogeny and classification of Lamiaceae subfamily Lamioideae. Taxon 60: 471-484.
Cao, Y.-N. Wang, I. J. Chen, L.-Y. Ding, Y.-Q. Liu, L.-X and Qiu, Y.- X. 2018. Inferring spatial patterns and drivers of population divergence of Neolitsea sericea (Lauraceae), based on molecular phylogeography and landscape genomics. Molecular Phylogenetics and Evolution 126: 162-172.
Cardoni, S. Piredda, R. Denk, T. Grimm, G. W. Papageorgiou, A. C. Schulze, E.-D. Scoppola, A. Shanjani, P. S. Suyama, Y. Tomaru, N. Worth, J. R. P and Simeone, M. C. 2022. 5S-IGS rDNA in wind-pollinated trees (Fagus L.) encapsulates 55 million years of reticulate evolution and hybrid origins of modern species. Plant Journal 109: 909-926.
Chen, D. Zhang, X. Kang, H. Sun, X. Yin, S. Du, H. Yamanaka, N. Gapare, W. Wu, H. X and Liu, C. 2012. Phylogeography of Quercus variabilis based on chloroplast DNA sequence in East Asia: Multiple glacial refugia and mainlandmigrated island populations. PLoS ONE 7: e47268.
Chen, Y.-P. Liu, A. Yu, X.-L and Xiang, C.-L. 2021. A preliminary phylogenetic study of Paraphlomis (Lamiaceae) based on molecular and morphological evidence. Plant Diversity 43: 206-215.
Chen, Y.-P. Sun, Z.-P. Xiao, J.-F. Yan, K.-J and Xiang, C.-L. 2022a. Paraphlomis longicalyx (Lamiaceae), a new species from the limestone area of Guangxi and Guizhou Provinces, southern China. Systematic Botany 47: 251-258.
Chen, Y.-P. Xiong, C. Zhou, H.-L. Chen, F and Xiang, C.-L. 2022b. Paraphlomis nana (Lamiaceae), a new species from Chongqing, China. Turkish Journal of Botany 46: 176-182.
Chung, G. Y. Jang, H.-D. Chang, K. S. Choi, H. J. Kim, Y.-S. Kim, H.-J and Son, D. C. 2023. A checklist of endemic plants on the Korean Peninsula II. Korean Journal of Plant Taxonomy 53: 79-101.
Corriveau, J. L and Coleman, A. W. 1988. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. American Journal of Botany 75: 1443-1458.
Guo, G.-X. Zhao, W.-Y. Chen, Y.-P. Xiao, J.-H. Li, Y.-Q and Fan, Q. 2023. Paraphlomis yingdeensis (Lamiaceae), a new species from Guangdong (China). PhytoKeys 219: 107-120.
Jin, D.-P. Lee, J.-H. Xu, B and Choi, B.-H. 2016. Phylogeography of East Asian Lespedeza buergeri (Fabaceae) based on chloroplast and nuclear ribosomal DNA sequence variations. Journal of Plant Research 129: 793-805.
Jin, J.-J. Yu, W.-B. Yang, J.-B. Song, Y. de Pamphilis, C. W. Yi, T.-S and Li, D.-Z. 2020. GetOrganelle: A fast and versatile tool-kit for accurate de novo assembly of organelle genomes. Genome Biology 21: 241.
Katoh, K and Standley, D. M. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30: 772-780.
Ko, S. C. Lee, Y.-M. Chung, K.-S. Son, D. C. Nam, B. M and Chung, G. Y. 2014. A new species of Paraphlomis (Lamiaceae) from Korea: An additional genus to the Korean flora. Phyto-taxa 175: 51-54.
Lee, J.-H. Lee, D.-H and Choi, B.-H. 2013. Phylogeography and genetic diversity of East Asian Neolitsea sericea (Lauraceae) based on variations in chloroplast DNA sequences. Journal of Plant Research 126: 193-202.
Li, X and Hedge, I. C. 1994. Lamiaceae. In Flora of China, Vol. 17. Verbenaceae through Solanaceae. Wu, Z.-Y. Raven, P. H (eds.), Science Press, Bejing and Missouri Botanical Garden, St, Louis, MO. 50-299.
Makino, T. 1915. Two new genera Matsumurella Makino and Ajugoides Makino. Botanical Magazine Tokyo 29: 279-283.
McCauley, D.E. 1995. The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends in Ecology and Evolution 10: 198-202.
Minh, B. Q. Schmidt, H. A. Chernomor, O. Schrempf, D. Woodhams, M. D. Von Haeseler, A and Lanfear, R. 2020. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530-1534.
Park, B. K. Kim, S.-C. Kim, Y.-S and Son, D. C. 2022. Complete chloroplast genome of Paraphlomis koreana (Lamiaceae): An endemic species from Korea. Journal of Asia-Pacific Biodiversity 15: 661-664.
Petit, R.J and Excoffier, L. 2009. Gene flow and species delimitation. Trends in Ecology and Evolution 24: 386-393.
Qiu, Y.-X. Fu, C.-X and Comes, H. P. 2011. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Molecular Phylogenetics and Evolution 59: 225-244.
Tagane, S and Nakanishi, H. 2021. Matsumurella chinensis (Lamiaceae), new to the flora of Japan. Journal of Japanese Botany 96: 175-179.
Xiang, C. Hu, G and Peng, H. 2016. New combinations and new synonyms in Lamiaceae from China. Biodiversity Science 24: 719-722.
Yuan, J.-C. Chen, Y.-P. Zhao, Y. Li, H.-B. Chen, B and Xiang, C.-L. 2022. Paraphlomis strictiflora (Lamioideae, Lamiaceae), a new species from Guizhou, China. Phytotaxa 575: 276-286.
Zhang, R.-B. Deng, T. Dou, Q.-L. Wei, R.-X. He, L. Ma, C.-B. Zhao, S and Hu, S. 2020. Paraphlomis kuankuoshuiensis (Lamiaceae), a new species from the limestone areas of northern Guizhou, China. PhytoKeys 139: 13-20.
|
|
||||||||||||||||||||||||||||||||||||||||||