PARK, JANG, SON, GIL, and KIM: Complete chloroplast genome sequence of Clematis calcicola (Ranunculaceae), a species endemic to Korea
Abstract
The complete chloroplast genome (cp genome) sequence of Clematis calcicola J. S. Kim (Ranunculaceae) is 159,655 bp in length. It consists of large (79,451 bp) and small (18,126 bp) single-copy regions and a pair of identical inverted repeats (31,039 bp). The genome contains 92 protein-coding genes, 36 transfer RNA genes, eight ribosomal RNA genes, and two pseudogenes. A phylogenetic analysis based on the cp genome of 19 taxa showed high similarity between our cp genome and data published for C. calcicola, which is recognized as a species endemic to the Korean Peninsula. The complete cp genome sequence of C. calcicola reported here provides important information for future phylogenetic and evolutionary studies of Ranunculaceae.
Keywords: Clematis, C. calcicola, Korean endemic species, phylogenetic relationships, plastome, simple sequence repeats
INTRODUCTION
The genus Clematis L. (Ranunculaceae) contains ca. 300 species of herbaceous or woody vines and a few erect shrubs and perennial grasses, which are common in temperate regions, including northern Europe, Siberia, and the Far East, but some species also occur in tropical regions ( Tamura, 1955, 1967, 1987, 1995; Wang and Bartholomew, 2001; Wang and Li, 2005; Kadota, 2006; Chang, 2007; Kim, 2017a; Chang and Kim, 2018; Park et al., 2022). Clematis calcicola J. S. Kim, is endemic to the Korean Peninsula ( Kim et al., 2009; National Institute of Biological Resources, 2011; Chung et al., 2017; Kim, 2017a, 2017b; Lee and Kim, 2018). This species is easily distinguishable from closely related taxa by its sparsely dentate, glabrous, and subcoriaceous leaflets and thick and smooth sepals ( Kim et al., 2009; Kim, 2017a). However, C. calcicola has limited distribution in Korea. In the present study, the complete chloroplast genome (chloroplast genome) of C. calcicola is reported for the first time, to the best of our knowledge. A phylogenetic analysis was also carried out to investigate the relationships of C. calcicola with other Clematis species. Chloroplasts are metabolic organelles with an important role in the physiology and development of terrestrial plants and algae ( Gray, 1989; Howe et al., 2003). The chloroplast genome is a suitable tool for studying the evolution and phylogeny of plants because of its highly conserved sequence and structure ( Maier et al., 1995). Chloroplasts have their own genetic replication mechanisms and they transcribe their genome relatively independently ( Fu et al., 2016). Most chloroplast genomes range 120 to 200 kb in length and exhibit a typical quadripartite structure, including a large single-copy sequence (LSC, 80–90 kb), a small single-copy sequence (SSC, 16–27 kb), and two inverted repeat sequences (IRs, 20–28 kb) with subequal length ( Palmer, 1985; Wang et al., 2008). The completion of cpDNA genome provides a large amount of information, including not only related information on protein-coding and non-coding genes but also data to infer gene rearrangement and evolutionary relationships ( Golenberg et al., 1993; Reith and Munholland, 1995). The complete chloroplast genome of C. calcicola, and the phylogenetic analysis results reported in the present study provide information on the relationship between C. calcicola and related species, which may be a valuable resource for future studies on this species.
MATERIALS AND METHODS
Clematis calcicola were sampled from Mt. Deokhang, Samcheok-si, Gangwon-do, South Korea (37o18′49.0″N, 129o00′41.5″E). The collected material was stored at the Herbarium of the Korea National Arboretum (KH contact, Dong Chan Son) under voucher Deokhangsan-190529-001.
Fresh leaves were silica-dried. Leaf DNA was extracted using a DNeasy Plant Mini Kit (Qiagen, Seoul, Korea) and verified on 2% agarose gel electrophoresis. The DNA library was constructed using the TruSeq Nano DNA Kit following the Sample Preparation Guide protocol provided by the manufacturer (Macrogen Inc., Seoul, Korea). Genome paired-end sequencing was performed at Macrogen Inc. on an Illumina platform (Illumina Inc., San Diego, CA, USA) based on 301 bp read size.
The complete chloroplast genome was assembled using Geneious v9.0.5 (Biomatters, Auckland, New Zealand) and annotated using the GeSeq tool ( Tillich et al., 2017) and Geneious v9.0.5 (Biomatters). To infer the phylogenetic relationships among Clematis species, the complete chloroplast genome sequences of 20 Clematis species were downloaded from GenBank ( https://www.ncbi.nlm.nih.gov/genbank/). Anemoclema glaucifolium (Franch.) W. T. Wang was used as the outgroup. Phylogenetic analysis was performed using 78 coding sequences of the Clematis species. Alignments were performed using MAFFT v7.450 ( Katoh et al., 2002; Katoh and Standley, 2013). A maximum likelihood (ML) bootstrap analysis with 1,000 replicates was performed and the best-fit model (transversion [TVM] + empirical base frequencies [F] + freeRate model parameters with two of categories [R2]) was determined using the IQ-tree web server ( Trifinopoulos et al., 2016). Bayesian inference (BI) analysis was conducted using MrBayes v3.2.6 ( Ronquist et al., 2012) in PhyloSuite ( Zhang et al., 2020). The best model of molecular evolution for the chloroplast genome dataset (general time reversible [GTR] + proportion of invariable sites [I] + rate of variation across sites [G] + F) was obtained in ModelFider v2.0 ( Kalyaanamoorthy et al., 2017).
RESULTS AND DISCUSSION
Clematis calcicola sequencing produced 6,657,552 reads, 7,417 of which corresponded to the chloroplast genome (depth 5.4). The complete chloroplast genome sequence of C. calcicola was 159,655 bp ( Fig. 1) and it was deposited in GenBank under accession number OK181902 and associated BioProject, SRA, and Bio-Sample numbers PRJNA886061, SRR21776408, and SAMN31117752, respectively. The LSC, SSC, and each of IRs of the chloroplast genome sequence were 79,451 bp, 18,126 bp, and 31,039 bp in length, respectively. The overall GC content of the chloroplast genome was 38%, and in the LSC, SSC, and each of IRs it was 36.3%, 31.3%, and 42%, respectively. The chloroplast genome contained 136 unique genes, including 92 coding genes, eight ribosomal RNA (rRNA) genes, 36 transfer RNA (tRNA) genes. Twenty-six genes (15 coding genes, four tRNA, and seven rRNA) were duplicated in the IRs. Also, infA identified as a pseudogene was included in the IRs ( Table 1).
We used sequences from 21 species (20 species of Clematis and one species of Anemoclema as an outgroup) to infer ML-based and BI-based phylogenies. Both phylogenies (ML and BI) had the same topology and showed high support for each branch ( Fig. 2). The phylogenetic analysis formed a monophyly of the genus Clematis. Nevertheless, it did not match the classification system divided into sections ( Wang and Li, 2005; Kim et al., 2009). However, C. calcicola and C. macropetala Ledeb. belonging to the section Atragene formed a group and was well supported (bootstrap value [BS] = 100; posterior probability [PP] = 1), forming a sister group to the section Cheiropsis (BS = 98; PP = 1), and branched first within the genus Clematis to form a basal group. We performed a comparative analysis with the chloroplast genome of C. macropetala, which was identified as the most closely related to C. calcicola in phylogenetic analysis ( Fig. 2). The chloroplast genome of C. calcicola was 11 bp longer than that of C. macropetala (159,647 bp, NC_041477). Nevertheless, C. calcicola and C. macropetala chloroplast genomes pairwise identity was 99.96%. Sixteen single nucleotide polymorphisms and 52 insertions/deletions (indels) were identified in the pairwise alignment of Clematis chloroplast genomes ( Tables 2 and 3). Of the 52 indels, 16 were found in four protein-coding regions ( clpP1, ndhF, ccsA, and ycf1) and 36 in the non-coding region between the two plastomes ( Table 3).
Clematis calcicola can be morphologically distinguished from C. macropetala by terete branches, ternate leaves, leaflet margins slightly dentate to dentate and subcoriaceous, and staminodes linear-spatulate to spatulate ( Kim et al., 2009; Yang et al., 2009). Although morphologically similar, the results of the present study support that C. calcicola is closely related but independent from C. macropetala, as evidenced by the genome differences and phylogenetic analysis. Also, C. macropetala is distributed in Central to Northeastern of China, E Mongolia, and Southeastern Russia, but C. calcicola is distributed limited in Korea.
Recently, three species of the Korean Clematis have been reported ( Choi et al., 2021; Park et al., 2021). The results of this study will provide important basic information for future phylogenetic and evolutionary studies of Clematis and Ranunculaceae by adding one species to them. In addition, comparative data between the two species within section Atragene of the genus Clematis will provide useful information for developing species identification markers and conducting genetic diversity analyses.
ACKNOWLEDGMENTS
This study was supported by the grant ‘Silvics of Korea’ [KNA-1-1-18, 15-3] financed by the Korea National Arboretum.
Fig. 1.
Gene map of Clematis calcicola complete chloroplast genome. Genes within the circle are transcribed in clockwise direction, and genes drawn out of the circle in counterclockwise direction. Different gene colors correspond to different gene functions. Inverted repeat (IR), small single-copy (SSC), and large single-copy (LSC) regions are indicated. 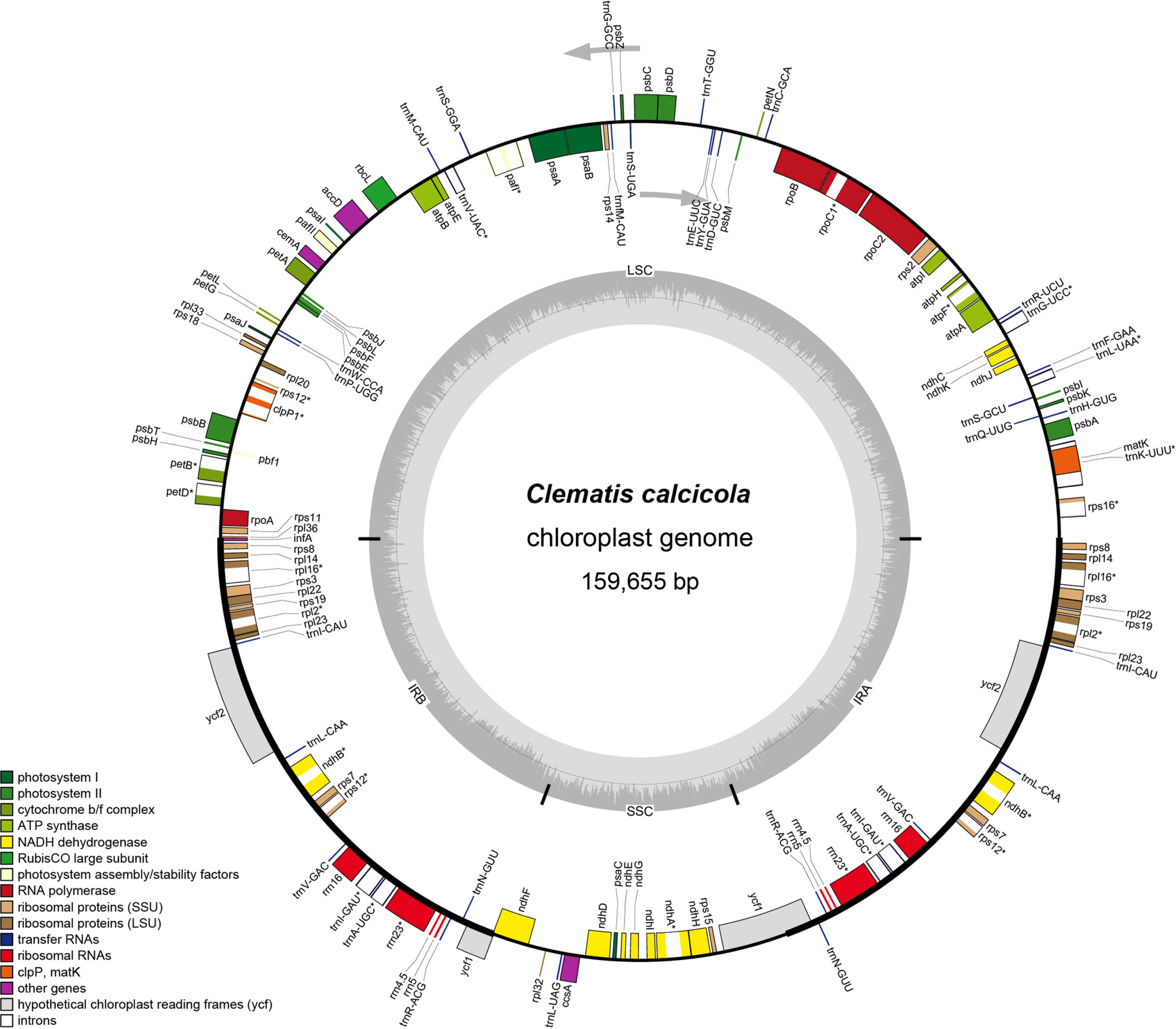
Fig. 2.
Phylogenetic tree of Clematis calcicola and related taxa based on 78 protein-coding gene sequences of chloroplast genome sequences using maximum likelihood (ML) and Bayesian inference (BI) analyses of different datasets. Numbers on each node are bootstrap values/posterior probability values. Anemoclema glaucifolium was set as the outgroup. 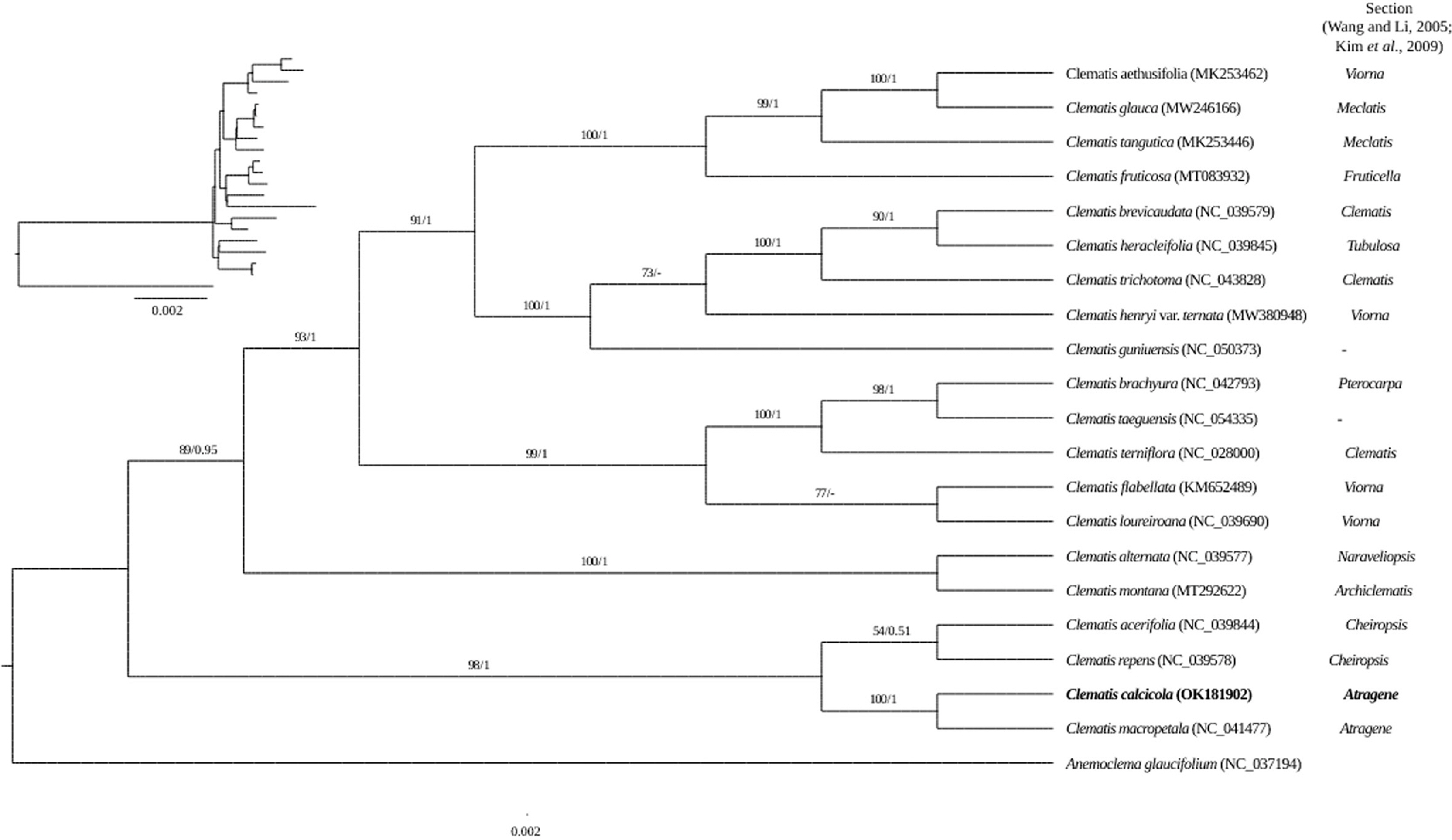
Table 1.
List of genes annotated in the chloroplast genome of Clematis calcicola.
|
Category for genes |
Group of genes |
Name of genes |
|
Self-replication |
Large subunit ribosomal proteins |
rpl2(×2)*, rpl14(×2), rpl16(×2)*, rpl20, rpl22(×2), rpl23(×2), rpl32, rpl33, rpl36
|
|
DNA-dependent RNA polymerase |
rpoA, rpoB, rpoC1*, rpoC2
|
|
Small subunit ribosomal proteins |
rps2, rps3(×2), rps4, rps7(×2), rps8(×2), rps11, rps12(×2)**T, rps14, rps15, rps16*, rps18, rps19(×2) |
|
Ribosomal RNAs |
rrn4.5S(×2), rrn5S(×2), rrn16S(× 2), rrn23S(×2) |
|
Transfer RNAs |
trnA-UGC(×2)*, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-GAU(×2)*, trnI-CAU(×2), trnK-UUU*, trnL-CAA(×2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(×2), trnP-UGG, trnQ-UUG, trnR-ACG(×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnV-GAC(×2), trnV-UAC*, trnW-CCA, trnY-GUA
|
|
Photosynthesis |
Subunits of ATP synthase |
atpA, atpB, atpE, atpF*, atpH, atpI
|
|
Subunits of NADH-dehydrogenase |
ndhA*, ndhB(×2)*, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK
|
|
Subunits of cytochrome b/f complex |
petA, petB*, petD*, petG, petL, petN
|
|
Subunits of photosystem I |
psaA, psaB, psaC, psaI, psaJ
|
|
Subunits of photosystem II |
psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbT, psbZ
|
|
Subunit of rubisco |
rbcL
|
|
Photosystem assembly factors |
pafI**, pafII
|
|
Photosystem biogenesis factor |
pbf1
|
|
Other genes |
Subunit of acetyl-CoA-carboxylase |
accD
|
|
C-type cytochrome synthesis gene |
ccsA
|
|
Envelop membrane protein |
cemA
|
|
ATP-dependent protease subunit P |
clpP**
|
|
Translational initiation factor |
ψinfA (×2) |
|
Maturase |
matK
|
|
Unknown function |
Conserved open reading frames |
ycf1, ycf2(×2) |
Table 2.
SNPs in Clematis calcicola relative to that of C. macropetala chloroplast genome.
|
No. |
C. calcicola
|
C. macropetala
|
Site |
Region |
Gene |
Position |
|
1 |
G |
A |
2,332 |
LSC |
rps16-trnK
|
IGS |
|
2 |
A |
C |
7,238 |
LSC |
psbA-trnH
|
IGS |
|
3 |
G |
A |
7,479 |
LSC |
trnQ-UUG
|
gene |
|
4 |
T |
C |
8,170 |
LSC |
psbK-psbI
|
IGS |
|
5 |
G |
A |
40,398 |
LSC |
trnS-psbZ
|
IGS |
|
6 |
A |
C |
70,138 |
LSC |
clpP1 intron |
intron |
|
7 |
T |
A |
79,713 |
IRB |
ψinfA-rps8
|
IGS |
|
8 |
C |
A |
110,507 |
SSC |
ndhF
|
gene |
|
9 |
T |
A |
110,523 |
SSC |
ndhF
|
gene |
|
10 |
A |
T |
115,484 |
SSC |
ccsA
|
gene |
|
11 |
T |
A |
119,716 |
SSC |
ndhG-ndhI
|
IGS |
|
12 |
A |
T |
119,774 |
SSC |
ndhG-ndhI
|
IGS |
|
13 |
T |
C |
119,775 |
SSC |
ndhG-ndhI
|
IGS |
|
14 |
T |
C |
119,786 |
SSC |
ndhG-ndhI
|
IGS |
|
15 |
C |
T |
119,787 |
SSC |
ndhG-ndhI
|
IGS |
|
16 |
A |
T |
159,421 |
IRA |
rps8-ψinfA
|
IGS |
Table 3.
Indels in Clematis calcicola relative to that of C. macropetala chloroplast genome.
|
No. |
C. calcicola
|
C. macropetala
|
Start |
End |
Length |
Region |
Position |
|
1 |
- |
T |
7,095 |
7,095 |
1 |
LSC |
psbA-trnH
|
|
2 |
- |
T |
10,568 |
10,568 |
1 |
LSC |
trnF-GAA-ndhJ
|
|
3 |
ATAT |
- |
12,739 |
12,742 |
4 |
LSC |
ndhC-trnG
|
|
4 |
A |
- |
35,889 |
35,889 |
1 |
LSC |
trnE-GCA-trnT
|
|
5 |
T |
- |
36,506 |
36,506 |
1 |
LSC |
trnT-GGU-psbD
|
|
6 |
A |
- |
37,116 |
37,116 |
1 |
LSC |
trnT-GGU-psbD
|
|
7 |
T |
- |
68,885 |
68,885 |
1 |
LSC |
rpl20-rps12
|
|
8 |
TTTATATT |
- |
70,561 |
70,568 |
8 |
LSC |
clpP1 intron |
|
9 |
- |
A |
75,155 |
75,155 |
1 |
LSC |
petB intron |
|
10 |
- |
A |
77,669 |
77,669 |
1 |
LSC |
petD-rpoA
|
|
11 |
A |
- |
110,499 |
110,499 |
1 |
SSC |
ndhF
|
|
12 |
T |
- |
113,545 |
113,545 |
1 |
SSC |
ndhF-rpl32
|
|
13 |
- |
A |
115,931 |
115,931 |
1 |
SSC |
ccsA
|
|
14 |
- |
TTTTTA |
116,163 |
116,168 |
6 |
SSC |
ccsA-ndhD
|
|
15 |
- |
AACTATCTA |
116,224 |
116,232 |
9 |
SSC |
ccsA-ndhD
|
|
16 |
T |
- |
118,371 |
118,371 |
1 |
SSC |
psaC-ndhE
|
|
17 |
- |
T |
118,831 |
118,831 |
1 |
SSC |
ndhE-ndhG
|
|
18 |
TCTA |
- |
119,779 |
119,782 |
4 |
SSC |
ndhG-ndhI
|
|
19 |
- |
A |
123,994 |
123,994 |
1 |
SSC |
ndhH-rps15
|
|
20 |
T |
- |
124,452 |
124,452 |
1 |
SSC |
rps15-ycf1
|
|
21 |
AATGAC |
- |
128,627 |
128,632 |
6 |
SSC |
ycf1
|
LITERATURE CITED
Chang, CS. 2007.
Clematis L. The Genera of Vascular Plants of Korea. Flora of Korea Editorial Committee (ed.), Academy Publishing Co, Seoul. 191-195.
Chang, CS and Kim, H. 2018.
Clematis L. The Genera of Vascular Plants of Korea. Flora of Korea Editorial Committee (ed.), Hongneung Science Publishing Co, Seoul. 253-259.
Choi, KS.. Ha, Y-H. Gil, H-Y. Choi, K. Kim, D-K and Oh, S-H. 2021. Two Korean endemic Clematis chloroplast genomes: inversion, reposition, expansion of the inverted repeat region, phylogenetic analysis, and nucleotide substitution rates. Plants 10: 397.    Chung, GY. Chang, KS. Chung, J-M. Choi, HJ. Paik, W-K and Hyun, J-O. 2017. A checklist of endemic plants on the Korean Peninsula. Korean Journal of Plant Taxonomy 47: 264-288.   Fu, P-C. Zhang, Y-Z. Geng, H-M and Chen, S-L. 2016. The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ 4: e2540.     Golenberg, EM. Clegg, MT. Durbin, ML. Doebley, J and Ma, DP. 1993. Evolution of a noncoding region of the chloroplast genome. Molecular Phylogenetics and Evolution 2: 52-64.   Gray, MW. 1989. The evolutionary origins of organelles. Trends in Genetics 5: 294-299.   Howe, CJ. Barbrook, AC. Koumandou, VL. Nisbet, RER. Symington, HA and Wightman, TF. 2003. Evolution of the chloroplast genome. Philosophical Transactions of the Royal Society of London Series B Biology Sciences 358: 99-107.   Kadota, Y. 2006.
Clematis L. In Flora of Japan. II: a. Angiospermae, Dicotyledoneae, Archichlamydeae. Iwatsuki, K. Boufford, DE. Ohba, H (eds.), Kodansha, Tokyo. 298-308.
Kim, J-S. Chung, J-M. Kim, S-Y and Park, J-H. 2009.
Clematis calcicola J. S. Kim: A new species of Clematis sect. Atragene (Ranunculaceae) from Korea. Korean Journal of Plant Taxonomy 39: 1-3.   Kim, JS. 2017a.
Clematis L. Flora of Korea. 2a: National Institute of Biological Resources (ed.), Incheon. 69-76.
Kim, M. 2017b. Korean Endemic Plants. Haejin Media Co. Ltd, Seoul. 76 pp.
Lee, JK and Kim, HS. 2018.
Clematis calcicola J. S. Kim. Silvics of Korea. 2: Oh, BU and Oh, SH. Korea National Arboretum of the Korea Forest Service, Pocheon. 63-71.
Maier, RM. Neckermann, K. Igloi, GL and Kössel, H. 1995. Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. Journal of Molecular Biology 251: 614-628.   National Institute of Biological Resources. 2011. Endemic Species of Korea. Geobook, Incheon. 331 pp.
Palmer, JD. 1985. Comparative organization of chloroplast genomes. Annual Review of Genetics 19: 325-354.   Park, BK. Ghimire, B. Ha, Y-H. Son, DC and Kim, D-K. 2021. Complete chloroplast genome of Clematis taeguensis (Ranunculaceae), an endemic species from South Korea. Mitochondrial DNA Part B Resources 6: 1496-1497.    Park, BK. Kim, J-S. Chung, GY. Kim, J-H. Son, DC and Jang, C- G. 2022.
Clematis pseudotubulosa (Ranunculaceae), a new species from Korea. Korean Journal of Plant Taxonomy 52: 35-44.   Reith, M and Munholland, J. 1995. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Molecular Biology Reporter 13: 333-335.   Ronquist, F. Teslenko, M. van der Mark, P. Ayres, DL. Darling, A. Höhna, S. Larget, B. Liu, L. Suchard, MA and Huelsenbeck, JP. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539-542.    Tamura, M. 1955. Systema clematidis Asiae Orientalis. Science Reports. 4: College of General Education, Osaka University, Osaka. 43-55.
Tamura, M. 1967. Morphology, Ecology and Phylogeny of the Ranunculaceae VII. Science Reports. 16: College of General Education, Osaka University, Osaka. 21-43.
Tamura, M. 1987. A classification of genus Clematis
. Acta Phytotaxonomica et Geobotanica 38: 33-44.
Tamura, M. 1995.
Clematis
. Die Natürlichen Pflanzenfamilien. Zweite Aufl. 17a. IV: Hiepko, P (ed.), Dunker & Humbolt, Berlin. 368-387.
Wang, WT and Bartholomew, B. 2001.
Clematis L. Flora of China. 6: Caryophyllaceae through Lardizabalaceae. Wu, ZY. Raven, PH. Hong, DY (eds.), Science Press, Beijing and Missouri Botanical Garden Press, St Louis, MO. 333-386.
Wang, W-T and Li, L-Q. 2005. A new system of classification of the genus Clematis (Ranunculaceae). Acta Phytotaxonomica Sinica 43: 431-488.
Yang, W-J. Li, L-Q and Xie, L. 2009. A revision of Clematis sect. Atragene (Ranunculaceae). Journal of Systematics and Evolution 47: 552-580. 
|
|













